Detection of bla-AIM Metallo Beta Lactamase Gene among Stenotrophomonas Maltophilia and Carbapenem Resistant Pseudomonas Aeruginosa Isolated from Various Infections in AL- Najaf Province
Enas Jalil Baqer AL-Mayali1, Sddiq Ghani Al-Muhanna2, Israa Abdul Ameer Al-Kraety3
1Jabir Ibn Hayyan Medical University, Iraq, Najaf, enas.j.baqir@jmu.edu.iq.
2,3Department of Medical Laboratory Technique, College of Health and Medical Techniques, University of Alkafeel Najaf, Iraq
* Correspondence: enas.j.baqir@jmu.edu.iq, Sddiq.g@gmail.com,
israa.ameer@alkafeel.edu.iq,
Available from. http://dx.doi.org/10.21931/BJ/2024.01.01.61
ABSTRACT
Stenotrophomonas maltophilia is a "rapidly evolving pathogen of concern" that is increasingly being identified. The World Health Organization also recognizes it as one of the hospitals' most significant multi-drug-resistant pathogens. Also, Pseudomonas aeruginosa is an opportunistic human pathogen that causes most healthcare-associated infections, and it is considered a paradigm of antibiotic resistance development. In many hospitals across the globe, carbapenem-resistant Pseudomonas aeruginosa has emerged as a significant source of infection. The present study aimed to study the isolation and diagnosis of S. maltophilia and P. aeruginosa from different clinical samples, evaluate the occurrence of carbapenem resistance of P. aeruginosa isolated from clinical samples, and investigate the dissemination of the bla-AIM genes between these isolates. Eight hundred fifty specimens were collected from various clinical samples between 2022 and 2023. The specimens included 220 swabs (burn), 200 (urine), 140 (stool), and 130(wound). 90 (ear),50 (throat), 10 (Cerebrospinal fluid), and 10 (blood). Represented by 680 specimens contained bacterial growth, and 170 specimens had no bacterial growth. Out of the 680 bacterial growth isolates, 410 revealed growths of Gram-negative bacteria, and 270 were Gram-positive bacteria. On MacConkey ag, ar 180/410 bacteria were lactose ferment; other isolates, es 230/410 of the isolates were lactose non-fermented bacteria. In a cross-sectional manner, Stenotrophomonas maltophilia and Pseudomonas aeruginosa isolates during this period were isolated and identified depending on the primary methods of diagnosis, then the use of the VITEK-2 compact system. The results showed 42 isolates of S. maltophilia and 80 isolates of P. aeruginosa from total Gram-negative bacteria. The results show that only five isolates contained the AIM gene, with a percentage of (10.4 %) of the 48 Carbapenem Resistant Pseudomonas aeruginosa isolates, five isolates from 42 S. maltophilia contain the AIM gene with a percentage (11.9%), based on the Polymerase chain reactions assay.
Keywords: Stenotrophomonas maltophilia, Carbapenem Resistance, Pseudomonas aerginosa.
INTRODUCTION
The opportunistic pathogen Stenotrophomonas maltophilia is becoming more critical. It has emerged as a hospital-acquired infection because of the widespread use of broad-spectrum antibiotics and the increasing number of invasive operations and immunocompromised patients. S. maltophilia exhibits a variety of antimicrobial resistance mechanisms, including the production of enzymes that hydrolyze or modify antibiotics, changes in membrane permeability, and a multi-drug efflux mechanism 1. Infections caused by P. aeruginosa, an opportunistic bacterium, include pneumonia, urinary tract infection, soft-tissue infection, and septicemia 2. Multidrug-resistant (MDR) P. aeruginosa infections have been linked to high rates of illness and death. Antibiotic treatment options are limited by the high degree of inherent and acquired resistance shown by P. aeruginosa. Carbapenems have been generally acknowledged as the most effective -lactams and extensively employed as the cornerstone and empirical therapy of severe infections caused by MDR P. aeruginosa; as a last-resort antibiotic, the discovery of carbapenem resistance is concerning, Carbapenem-resistant P. aeruginosa has now been discovered and is spreading across the globe 3. When it comes to the hydrolysis of carbapenem and other beta-lactamases (excluding monobactam), metallo beta-lactamases have a high degree of efficacy and are unaffected by any of the currently known clinically available beta lactamases-inhibitors, all but a few MBL genes are found in integron gene cassettes coupled to mobile elements, which makes it easier for them to be transferred across different bacterial genera and species through horizontal gene transfer 4. In the early 1990s, the Impeneme and Vancomycin-type enzymes, the most prominent of the acquired Metalobeta-lactamase, were discovered, and since then, several more kinds of acquired Metalobeta-lactamase enzymes 5. This study aims to determine the occurrence of the bla-AIM gene among multi-resistance S. maltophilia and P. aeruginosa isolated from hospitals in Najaf.
MATERIALS AND METHODS
Patients Demography
Between 2022 and 2023, 850 clinical samples were taken from patients at the Al-Sadder Medical City, Al-Hakeem General Hospital, Al-Forat General Hospital, Al-Zahra Maternity and Children, Al-Sajad Hospital, and Burn Center in the province of Al-Najaf who were afflicted with various infections. Al-Sadder Medical City 190 (22.3%) and Burn Center 220 (25.8%) produced the majority of the isolates, respectively, as shown in Table (1).
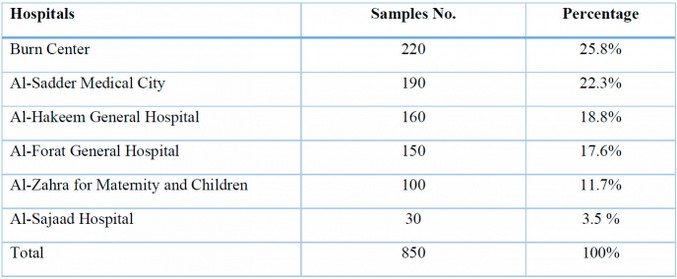
Table (1): Distribution of the number of clinical samples in different Najaf hospitals
All samples were cultured on MacConkey medium and blood agar. After a 24-hour incubation period, the results showed 680 samples containing only bacterial growth. Two hundred two isolates represented the bacterial growth isolates were recovered from burn infections, 178 from urinary tract infections, 100 were from gastroenteritis, 103 bacterial isolates recovered from wounds, and 60, 30, and 3,4 were obtained from ear, throat, cerebrospinal fluid, blood respectively as show Table (2 ).
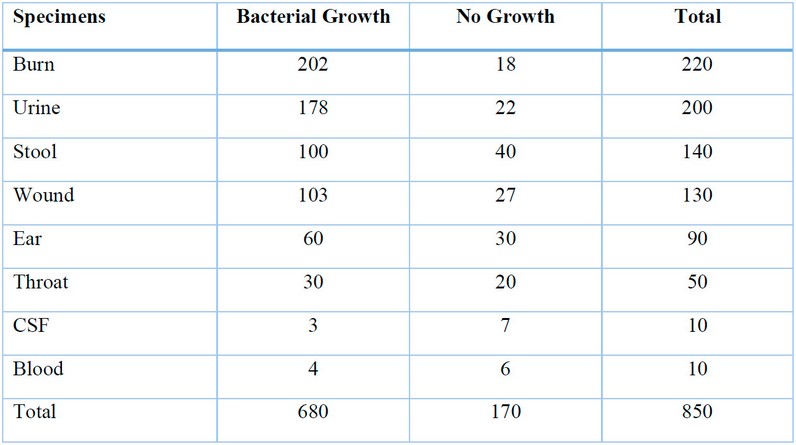
Table (2): Distribution of bacterial growth with infection site
Isolation and Identification of Stenotrophomonas maltophilia and Pseudomonas aerginosa Isolates.
The colony appearance, microscopic inspection, and biochemical characteristics were used to identify bacterial isolates. 410 of the 680 bacterial growth isolates showed Gram-negative bacteria growth, while the remaining 270 could not grow on the differential medium (Mac-Conkey Agar) utilized in this investigation. On MacConkey agar, 180/410 of the bacteria produced pink colonies since the bacteria lactose fermented grew on MaCconkey agar and produced pink colonies; other isolates 230/410 isolates produced yellow or colorless colonies since they were lactose non-fermented bacteria. In microscopic examination (Gram film), the organism appeared as a Gram-negative bacillus with a slightly smaller size. These are 230 samples, on which many of the biochemical tests available were conducted, including catalase, oxidase, motility, IMVICs, urease and TSI test to approximate the results for the diagnosis of Stenotrophomonas maltophilia and Pseudomonas aeruginosa using VITEK2 system. The results showed the presence of 230 Gram-negative bacterial isolates that did not ferment to lactose after growing on MacConkey, including 42 isolates of S. maltophilia and 80 isolates of P. aeruginosa.
As a result of S. maltophilia having polar flagella, it became mobile and grew effectively on MacConkey, in addition to producing a distinctive pigment on the medium. The catalase test was positive, and the oxidase test was negative, distinguishing it from other members of the genus. In addition, the IMVC test was negative, except for the citrate test, the result of which was different between the isolat, es, but P. aeruginosa produces pyocyanin and gives off a strong oxidase positive7.
Antibiotic susceptibility test to detect carbapenem resistance P. aeruginosa.
The Eighty clinical isolates of P. aeruginosa were examined for their ability to develop in the presence of carbapenem drugs, Using the Kirby-Bauer disk diffusion technique and antipseudomonal drugs (imipenem and meropenem) from one antimicrobial category (be-ta-lactam medication), recommendations were followed to conduct preliminary susceptibility screening on the isolates. The findings of drug susceptibility testing include 48 isolate resistance to imipenem and 44 isolate resistances to meropenem 8.
DNA extraction
Based on the commercial kits used to extract the genome, the kit (Favorgen, Taiwan) was used in this study.
Molecular identification
The bla-AIM gene for S. maltophilia and P. aeruginosa was detected by PCR using a specific primer with the sequence mentioned in Table 3. Amplification conditions were performed as previously described by (Reference) and summarized in Table 4

Table (3). Primer used in this study
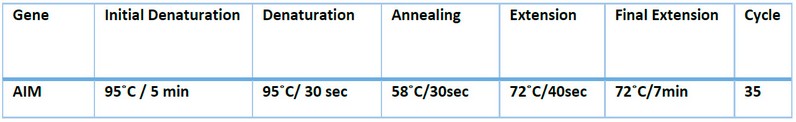
Table (4). PCR program to (bla-AIM gene).
RESULTS AND DISCUSSION
Molecular screening of bla-AIM producers S. maltophilia.
Using specialized Ambler class B MB (AIM) primers, all S. maltophilia isolates underwent conventional PCR screening for possible MBL gene determinants. With only five isolated positive findings and a proportion of (11.9%) %) as shown in Figure (1).
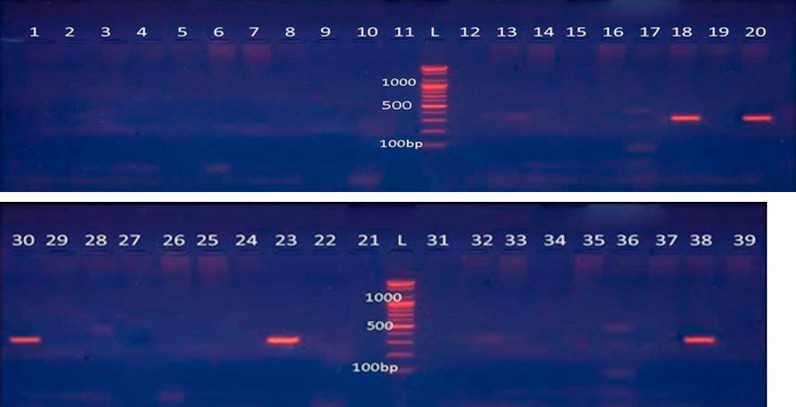
Figure 1: Amplification of bla-AIM gene by PCR from S. maltophilia isolates. Lane (18, 20, 23, 30, 38) shows positive results of the bla-AIM gene with 322 bp.
Even though multiple stable medications and inhibitor combinations are in different phases of development, they elude all recently approved -lactam—lactamase inhibitor combinations.9 in Iraq reported that S. maltophilia is an emerging opportunistic nosocomial pathogen causing various infections. Ceftazidime and chloramphenicol did not affect any of the specimens. Moreover, 100 percent and 43 percent of them developed extended spectrum β-lactamases (ESBLs) and carbapenemases, respectively.
Molecular screening of bla-AIM producers to carbapenem resistance Pseudomonas aeruginosa.
All 48 carbapenem-resistant P. aeruginosa isolates were screened by conventional PCR for potential gene determinants encoding MBL using specific Ambler class B MBL(AIM) primers. Only five isolates had positive results for this gene with a percentage (10.4%), as shown in Figure (2).
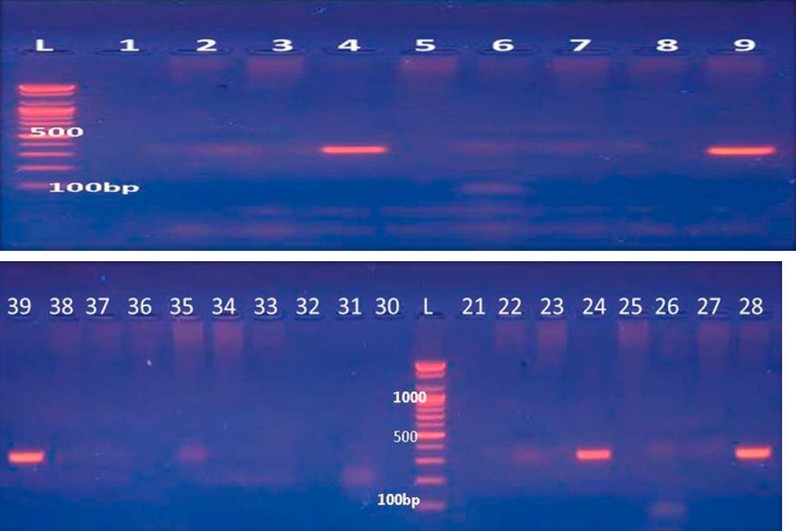
Figure 2. Amplification of bla-AIM gene by PCR from Pseudomonas aeruginosa isolates. lanes (4, 9, 24, 38, 39) show positive results of the bla-AIM gene with 322 bp.
S. maltophilia has shown high resistance to many antibiotics, including beta-lactams, aminoglycosides, and chloramphenicol because it contains genes carried on the chromosome such as blaL1 metallo β-lactamase and blaL2 β-lactamase genes, this was consistent with the isolate's resistance profile10. This result agrees with another study in Iraq showing that S. maltophilia possesses many resistance genes, the most important of which is the metallo beta-lactamase gene11.
Contrary to the other B3 MBLs (such as Stenotrophomonas maltophilia L1 and Janthina bacteria lividum THIN-B, Chryseobacterium meningosepticum GOB, Legionella gormanii FEZ-1, and Caulobacter crescentus CAU-1), this one can survive in the presence of antibiotics. Despite having the same MBL fold, B3 enzymes have a significantly different active site architecture than other subclasses 12.
An earlier investigation found that Carbapenemase synthesis in P. aeruginosa is particularly critical because of the fast spread of CRPA due to the acquisition of carbapenemase genes through mobile genetic elements. P. aeruginosa has been shown to generate carbapenemases of classes A, B, and D so far, with the most common being the Verona Integron-encoded Metallo—lactamase (VIM), imipenemases (IMP), and New Delhi Metallo—lactamase (NDM) all belonging to class B Metallo—lactamase (MBL) enzymes 13
This result also agrees with another research study, P. aeruginosa, a prominent human pathogen, which produced AIM-1. This subclass B3 enzyme was the first to be discovered on a mobile genetic element; AIM-1 hydrolyzes most lactams, except aztreonam and ceftazidime, to a lesser extent. However, it has much higher activity for cefepime and carbapenems than most other MBLs14.
CONCLUSIONS
The presence of isolates of S. maltophilia, causing several infections, containing a gene (bla-AIM) with a percentage (11.9%). Also, Carbapenem Resistant Pseudomonas aeruginosa isolates caused several infections, containing a gene (bla-AIM) with a percentage (10.4%).
Author Contributions: “Conceptualization, E.J.B.A. and I.A.A.; methodology, E.J.B.A. and S.G.A.; software,E.J.B.A..; validation, S.G.A., I.A.A and E.J.B.A.; formal analysis, I.A.A.; investigation, E.J.B.A..; resources, S.G.A.; data curation, I.A.A and S.G.A.; writing—original draft preparation, E.J.B.A.; writ-ing—review and editing, I.A.A.; visualization, I.A.A.; supervision, E.J.B.A.; project administration, I.A.A.; funding acquisition, S.G.A., I.A.A and EJBA. All authors have read and agreed to the published version of the manuscript."
Funding: "This research received no external funding".
Informed Consent Statement: Not applicable
Acknowledgments: The authors would like to express thanks and appreciation to the presidency of the University of Alkafeel for the support in completing this research.
Conflicts of Interest: "The authors declare no conflict of interest."
REFERENCES
1. Huang Y.-W., Liou R.-S., Lin Y.-T., Huang H.-H., and Yang T.-C.A linkage between SmeIJK efflux pump, cell envelope integrity, and σEmediated envelope stress response in Stenotrophomonas maltophilia.2014.
2. Rosenthal V.D., Bijie H. Maki D.G., Mehta, Y. Apisarnthanarak A. and Medeiros E.A. International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for2004–2009. Am J Infect Control .2012;40:396–407.
3. Vural E., Delialioglu N., Ulger S. T., Emekdas G. and Serin M. S. Phenotypic and molecular detection of the metallo-beta-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from clinical samples. Jundishapur, J. Microbiol. 2020 e90034: 1-8. 10.5812/jjm.90034.
4. Edelstein M.V., Skleenova E.N., Shevchenko O.V., D'Souza J.W., Tapalski D.V., Azizov I.S., Sukhorukova M.V., Pavlukov R.A., Kozlov R.S., Toleman M.A. and Walsh T.R. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis. 2013; 13:867-76.
5. Wachino J., Yoshida H., Yamane K., Suzuki S., Matsui M., Yamagishi T., Tsutsui A., Konda T., Shibayama K. and Arakawa Y. SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem- resistant Serratia marcescens clinical isolate. Antimicrob Agents Chemother.2011 ;55:5143-9.
6. Miller, R. D., Iinishi, A., Modaresi, S. M., Yoo, B. K., Curtis, T. D., Lariviere, P. J., ... & Lewis, K. (2022). Computational identification of a systemic antibiotic for gram-negative bacteria. Nature Microbiology, 7(10), 1661-1672.
7. Koneman E. W., Allen S. D., Janda W.M., Winn W.C., Procop G.W., Schreckenberger P.C. and Woods,G.L.The non fermentive gram negative bacilli, Color Atlas And Textbook of Diagnostic Microbiology, 6 th edition, (J.B.Lippincott Co., Philadelphia, ).2006;316-328.
8. Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs in context. 2018;7.
9. Poirel L., Walsh T. R., Cuvillier V. and Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagnostic microbiology and infectious disease.2011; 70(1), 119-123.
10. Kumwenda, G. P.; Kasambara, W.; Phiri, A.; Chizani, K.; Banda, A.; Choonara, F. and Lichapa, B. (2021). A multidrug-resistant Stenotrophomonas maltophilia clinical isolate from Kamuzu Central Hospital, Malawi. Malawi Medical Journal, 33(2), 82-84.
11. Saleh, R. O.; Hussen, B. M.; Mubarak, S. M. and Mostafavi, S. K. (2021). High diversity of virulent and multidrug-resistant Stenotrophomonas maltophilia in Iraq. Gene Reports, 23.
12. Khorvash F., Yazdani M., Shabani S. and Soudi A. Pseudomonas aeruginosa-producing metallo-β-lactamases (VIM, IMP, SME, and AIM) in the clinical isolates of intensive care units, a university hospital in Isfahan, Iran. Advanced biomedical research.2017;6.
13. Bording-Jorgensen M., Tyrrell H., Lloyd C., & Chui, L. Comparison of Common Enrichment Broths Used in Diagnostic Laboratories for ShigaToxin—Producing Escherichia coli. Microorganisms.2021; 9(3), 503.
14. Yong, D.; Toleman, M. A.; Bell, J.; Ritchie, B.; Pratt, R.; Ryley, H. and Walsh, T. R. (2012). Genetic and biochemical characterization of an acquired subgroup B3 metallo-β-lactamase gene, bla AIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrobial agents and chemotherapy, 56(12), 6154-6159.
Received: October 9th 2023/ Accepted: January 15th 2024 / Published:15 February 2024
Citation: AL-Mayali E. J. B., Al-Muhanna S. G., Al-Kraety I. A. A. Detection of bla-AIM Metallo Beta Lactamase Gene among Stenotrophomonas maltophilia and Carbapenem Resistant Pseudomonas Aeruginosa Isolated from Various Infections in AL- Najaf Province. Revis Bionatura 2024; 1 (1) 61. http://dx.doi.org/10.21931/BJ/2024.01.01.61
Additional information Correspondence should be addressed to enas.j.baqir@jmu.edu.iq
Peer review information. Bionatura thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).

