Preliminary Assessment of the In Vitro Immunological Activity of Native Lactobacillus Strains from the Ecuadorian Amazon
Laura Scalvenzi 1*, Roldán Torres Gutiérrez 2
1 Universidad Estatal Amazónica / Puyo /Ecuador;
2 Universidad Regional Amazónica IKIAM / Tena / Ecuador;
3 Universidad Técnica de Ambato / Ambato / Ecuador;
* Correspondence: lscalvenzi@uea.edu.ec
ABSTRACT
The isolation and characterization of new Lactobacillus strains from fermented traditional foods is a global trend as it enhances the potential for discovering novel probiotic foods. The fermented cocoa (Theobroma cacao) mucilage is an intriguing substrate for isolating lactic acid bacteria involved in fermentation. This study represents a preliminary investigation into the isolation, quantification, characterization, and immunological activity of Lactobacillus strains derived from cocoa beans sampled from two farmer associations (Kallari and Wiñak) in the Ecuadorian Amazon region. A mother culture was prepared using fermented cocoa pulp, cultivated on selective MRS media. After growth, the isolates were morphologically characterized. A significantly higher bacterial concentration was recorded in Kallari Association samples if compared with Wiñac. A total of 25 strains were isolated, eight of which were rod-shaped and positive to catalase tests and were characterized as Lactobacillus. In vitro, immunological activity was performed on differentiated THP-1 cell lines. Cells were treated with bacterial concentrates, and immunological activity was determined through interleukin-10 expression. Results indicated that W6 strain showed the highest immunological activity. These results indicated that Lactobacillus strains isolated from fermented cocoa pulp in the Ecuadorian Amazon show promise as a new source of probiotics.
Keywords: cocoa; lactic acid bacteria; biological activity; isolation; cocoa fermentation; probiotics; cocoa pulp
INTRODUCTION
The human body balances gastrointestinal microbiota and the immune system 1,2. Disruption of this microbiota can lead to diseases, discomfort during food consumption, and difficulties in nutrient absorption3. Probiotics and derivatives of beneficial bacteria help promote immune system balance by inducing the secretion of anti-inflammatory cytokines and other factors that reduce non-specific immune responses and the reactivity of macrophages and lymphocytes 4,5. Bacteria from the Lactobacillus genus and others have been shown to support gastrointestinal homeostasis 6. Many Lactobacillus species can thrive during the fermentation of fruit beverages. Evidence suggests that once isolated and tested in vitro, in vivo, and during human consumption, these bacteria could favorably influence the microbiota and immune system equilibrium 7,8.
The isolation and characterization of novel Lactobacillus strains from traditionally fermented foods is a rapidly expanding field worldwide. This approach holds significant potential for the development of new probiotic products using native bacterial strains that are specifically adapted to local conditions 9-11.
Among such fermented foods, cocoa (Theobroma cacao) mucilage is a relevant growth substrate for bacteria with potential probiotic properties. The cocoa plant is native to Ecuador, as evidenced by archeological findings of theobromine alkaloid in the oldest organic cocoa matter discovered in Zamora Chinchipe province 12. Today, cocoa cultivation remains a cornerstone of economic sustainability for Amazonian communities in Ecuador, where, for many, the sale of cocoa beans is the primary source of income 13. Furthermore, cocoa cultivation is critical in protecting the rainforest against slash-and-burn practices, a significant driver of agricultural expansion that threatens the fragile Amazonian ecosystem. While factors such as genotype, kernel composition, soil type, and plantation age influence the development of cocoa flavors 14, post-harvest practices like bean fermentation are essential for reaching the quality standards demanded by the market 15.
The specific microbial inoculum determines the flavor compounds generated during cocoa fermentation. These microbes drive the biochemical processes that form free amino acids, peptides, and sugars at the end of fermentation, ultimately shaping the desired quality of fermented cocoa beans 12,16. Lactic acid bacteria (LAB), particularly species belonging to the genus Lactobacillus, are essential for achieving optimal results. These bacteria contribute to cocoa flavor development and hold potential as a foundation for functional probiotic foods 17, potentially enhancing the value of Amazonian cocoa products. To our knowledge, a significant gap exists in understanding Ecuador's microbial communities involved in cocoa bean fermentation. Although these local microorganisms have the potential to act as probiotics, their applications remain largely unexplored. Delving into this understudied niche offers a valuable opportunity to impact the Ecuadorian cocoa industry positively.
The isolation, characterization, and evaluation of these microorganisms could lead to the development of novel fermentation techniques that enhance the quality of Ecuadorian chocolate while introducing health benefits. Therefore, studies on native Lactobacillus strains represent a significant opportunity for the Ecuadorian Amazon. If these native microorganisms exhibit probiotic properties, they could offer multiple advantages, including producing new functional foods, increased economic opportunities for local communities, and potential improvement in the immune system of the region's inhabitants, who face the highest malnutrition rates in the country 18.
No studies have been reported to elucidate the immunological function of native LAB strains or species with probiotic potential in the Ecuadorian Amazon region. This is why this preliminary research becomes the first approach to the isolation, characterization, and determination of initial immunological properties of bacteria of the Lactobacillus genus from fermentation processes of cocoa seeds collected from Napo, Ecuador.
MATERIALS AND METHODS
Sampling, isolation, and counting of Lactic Acid Bacteria (LAB)
Cocoa bean samples were collected from fermentation boxes belonging to the Kallari (K) and Wiñak (W) farmer associations in Napo province, Ecuador. The microbial communities involved in the fermentation of cocoa mucilage in all Amazonian regions of Ecuador, including Napo, remain largely unexplored. This study aims to address this knowledge gap by investigating the microbial ecology of cocoa fermentation in the Napo region. Given the presence of well-established cacao producer associations, Kallari and Wiñiak, with the necessary infrastructure and expertise, their involvement was crucial to ensure the successful collection and analysis of samples.
Yeasts dominate the initial stages of cacao mucilage fermentation. Their growth creates optimal conditions (anaerobic environment) for the development of LAB, which begins to proliferate significantly within 24-72 hours 19, 20, 21. For this reason, samples for this study were collected on the second day of fermentation. Four samples (each 200 grams) were taken from each fermentation box. Each sample consisted of five subsamples collected at a depth of 30 cm from five different locations within box 19. Samples were assigned alphanumeric codes: "K" or "W" for Kallari or Wiñak association, respectively, and "S" followed by a number for the subsample (e.g., W1S1, W1S2, W2S1, W2S2, K1S1, K1S2, K2S1, K2S2). Seeds were stored in airtight zip-lock bags and refrigerated at 4°C for transport and further laboratory processing.
Isolation of LAB was carried out following the method described by Camu et al. 22. Twenty grams of cocoa seeds, from each sample, were mixed with 180 ml of peptone water and shaken vigorously for 3 minutes to create a homogenous solution containing pulp particles. One milliliter of this solution was used for serial dilutions. Aliquots (0.1 ml) containing an estimated 105 to 107 CFU were inoculated into three Petri dishes containing Man-Rogosa-Sharpe (MRS) agar. The plates were incubated for 72h at 37°C to observe colony growth.
Characterization of Lactobacillus strains
After incubation, colony-forming units were counted for each sample and dilution. Biochemical tests such as Gram stain, catalase test, and growth parameters (color, shape, elevation, borders) were used to identify the genus. Only colonies exhibiting morphological characteristics consistent with the genus Lactobacillus, such as rod-shaped, Gram-positive, non-pigmented, catalase-negative, and anaerobic, were considered 23. Catalase test was carried out following MacFaddin's method 24. Each strain was transferred to a slide, and a drop of 3% hydrogen peroxide was added. The reaction was visualized under a 40x magnifying optical microscope. The presence or absence of bubbles indicates catalase activity: no bubbles indicate a negative reaction (no catalase enzyme to break down hydrogen peroxide), while bubble formation indicates a positive reaction. Despite advancements in molecular techniques, biochemical tests remain a cornerstone of taxonomic identification for the genus Lactobacillus 25, 26, 27.
Each strain was assigned a unique alphanumeric code consisting of a letter representing the farmer association ("K" for Kallari or "W" for Wiñak) followed by a number denoting the isolated strain. Finally, all isolates displaying morpho-cultural characteristics of LAB were cryopreserved in 50% (v/v) glycerol solution.
In vitro immunological activity of Lactobacillus
LAB isolated from cocoa mucilage that presented morphological and biochemical characteristics belonging to the genus Lactobacillus (Gram + bacilli, catalase-negative) were studied to determine their immunological activity. The assay was performed on the human acute leukemia monocyte cell line THP-1 from the American Type Culture Collection (ATCC) at different stages of differentiation (M0, M1, M2). The THP-1 cell line is one of the most used models to test the immune regulatory properties of probiotics derivate from bacteria 1,2,6 . THP-1 cells were transformed into macrophages by activation using PMA (Phorbol 12-myristate 13-acetate). Once differentiated into an M0 macrophage state, they are incubated with IL-3 and IL-4 to obtain M2 macrophages or with IFN-gamma and lipopolysaccharide (LPS) for macrophage activation (M1) 28. In this study, 106 cells/ml concentration was seeded in six-well plates and exposed to 50 nm phorbol myristate acetate (PMA) to induce differentiation for 24 h. Cells were then washed and exposed to a wash water bath. Subsequently, cells were maintained without stimuli for 24 h and then subjected to the effect of the bacteria under study. The assay was duplicated using 1 mg/ml pellet of the bacterial strains. The expression of interleukin-10 (IL10) and tumor necrosis factors (TNFα), measured by qPCR and ELISA, was evaluated in differentiated THP-116 macrophages.
Statistical analysis
A comparison of means for data obtained from bacterial quantification was processed using the SPSS v. 24 statistical package. Simple ANOVA with Tukey HSD test and significance level p<0.05 was used for both inter-sample and inter-locality differences.
Statistical analysis of the immunological activity data was performed using GraphPad Prism 6. A Kolmogorov-Smirnov normality test was conducted to determine the use of parametric or non-parametric analysis. Then, non-parametric ANOVA and Mann-Whitney rank test were used to analyze differences between conditions, using an alpha value of 0.05 (* p <0.05, ** p <0.01).
RESULTS
Isolation and quantification of LAB
Figure 1 presents the quantification of LAB isolates following a 72-hour incubation period on Petri dishes. Analysis of LAB quantification revealed a more uniform distribution of colony-forming units (CFUs) in samples collected from Wiñak (panel A), with no significant differences observed. In contrast, Kallari samples (panel B) displayed a higher overall CFU count than Wiñak, and considerable variations were evident between the collected subsamples. This finding underscores the relevance of collecting and analyzing composite samples, to ensure a representative picture of the LAB population within different fermentation boxes at each sampling site. Mixed bacterial cultures were observed in the samples analyzed (Figure 2).
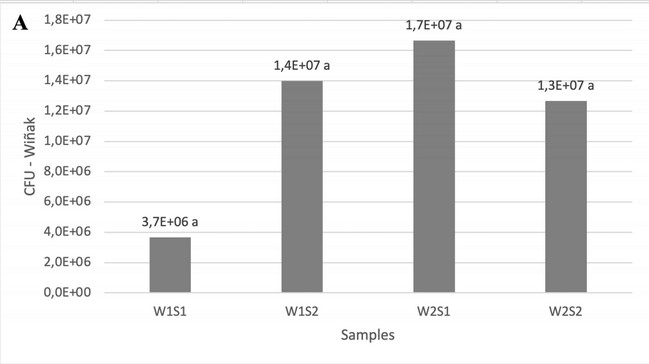


Figure 1. Bacterial quantification in fermenting cocoa bean samples from Wiñak (A) and Kallari (B) and the average of samples in both locations (C). Different letters in the bars represent significant differences p<0.05, Tukey HSD.

Figure 2. Mixed culture of LAB on MRS agar, isolated from mother cultures of fermenting cocoa beans from Kallari association
Characterization of Lactobacillus strains
Bacterial isolates exhibiting morpho-cultural characteristics consistent with LAB were selected for further characterization. Table 1 presents the 25 isolates identified that showed distinct morpho-cultural features.

1Color: 1-yellow, 2-light yellow, 3-opaque, 4-white, 5-pinkish; 2Elevation: C-convex, P-flat; 3Margin: E-entire, I-irregular; 4Consistency: -none slime, +light slime, ++moderate slime, +++ abundant slime; 5Growth: -none, +light, ++moderate, +++ abundant; 6Gram: -negative, +positive; 7Catalase: -negative, +positive.
Table 1. Morpho-cultural and biochemical characteristics of bacteria isolated from cocoa bean mucilage at Kallari and Wiñak associations
Gram staining, microscopic observation, and catalase test allowed the identification of eight isolates (W1, W2, W4, W6, W7, K6, K10, K13) showing a bacillus shape, belonging to the genus Lactobacillus (Table 1, Figure 3).

Figure 3. Lactobacillus spp. Observed at optical microscope, isolated from Kallari sample
(see black arrows).
Preliminary in vitro immunological activity of Lactobacillus isolates
To assess the immunological activity of Lactobacillus isolates, they were co-incubated with TPH-1 cell lines. Figure 4 illustrates the effects of different strains on TNFα and IL-10 expression. Panel B shows that W6 strain revealed the highest significant levels of IL-10 secretion by macrophages as measured by ELISA. This finding was further corroborated by the qPCR analysis of IL-10 expression (panel D).

Figure 4. Immunological activity of Lactobacillus isolates. A: TNFα expression by ELISA; B: IL-10 expression by ELISA; C: TNF expression by qPCR; D: IL-10 expression by qPCR.
DISCUSSION
Temperature, incubation time, and concentration of acetic and lactic acids affect the cocoa fermentation process. Those factors highly influence the production of metabolites essential for developing aroma, flavor, and high-quality chocolate precursors 12,29,30. The LAB populations within fermentation boxes revealed differences in bacterial concentration, depending on the geographical proceeding of the cocoa bean sample. As reported in other studies, this situation suggests adapting the microorganisms to the environment they originated in 31.
The analysis of the samples showed the presence of mixed microbial cultures, as other studies showed. Cocoa fermentation is a spontaneous process that naturally occurs thanks to microbes already present in the environment. These microbes feed on the bean pulp, reducing oxygen and creating ideal conditions for them to thrive. Several microbes include yeasts, lactic acid, and acetic bacteria.
The initial fermentation stage is characterized by a reduction in oxygen diffusion within the bean matrix due to the surrounding pulp volume, which creates an anaerobic environment. During this phase, a sequential microbial dominance occurs. Yeasts are first developed by consuming sugars and producing ethanol. The alcoholic fermentation produces an alkaline environment that, together with alcohol and oxygen, inhibits yeast activity and prevents lactic bacteria development. In the second fermentation phase, more oxygen enters the matrix, creating adequate conditions for lactic bacteria growth that converts pulp sugars into lactic acid.
Unlike other fermented foods, cocoa relies heavily on the beans' enzymes to develop flavor. Without fermentation, cocoa beans would be tasteless. Microbes break down the pulp and create essential flavor compounds 12,19 during this process.
The highest LAB concentration is typically recorded on the second day of fermentation, including for the Lactobacillus genus. Gram staining, microscopy, and the catalase test confirmed the presence of bacteria with bacillary morphology, characteristic to Lactobacillus genus. These results align with those reported by previous research 12,30,32,33. While recent studies increasingly employ molecular analyses for specific LAB strain identification, several authors suggest that morphological, cultural, and biochemical characterization is enough to determine Lactobacillus presence in various samples 23,34,35.
The W6 strain recorded the highest significant levels of IL-10 secretion by macrophages. THP-1 differentiated macrophages generally exhibit a pro-inflammatory signature characterized by high TNFα expression under controlled conditions 4,36. However, according to some authors, IL-10 expression is the most crucial indicator of an anti-inflammatory and homeostatic macrophage lineage, a key characteristic of probiotics 37. The ability of the W6 strain to stimulate IL-10 secretion suggests its potential probiotic properties.
CONCLUSIONS
Given the recent discovery that the cocoa plant is endemic to the Ecuadorian Amazon 38, research into this species has increased importance for local economic development and future studies that could lead to additional quality certifications. This study is distinguished by its focus on the probiotic potential of mucilage derived from cocoa cultivated in the region of Napo, located in Amazonian Ecuador.
Analysis of LAB isolates from cocoa mucilage samples collected in Napo, under the fermentation process, revealed a rich diversity based on morphology, culture, and biochemistry. Kallari samples displayed higher LAB concentrations, potentially influencing cocoa bean quality and processing. Furthermore, in vitro immunological studies identified W6 strain as the most promising candidate due to its ability to induce IL-10 secretion in macrophages, suggesting its potential probiotic properties. In vitro experiments were decided to provide preliminary scientific results for future research. Data obtained in the present study provide sufficient justification to carry on with this research, focusing on (i) performing assays on in vivo models, (ii) studying other types of anti-inflammatory cytokines, as well as (iii) the evaluation of additional probiotic activities as survivability under simulated gastrointestinal conditions, adhesion to intestinal epithelial cells and antimicrobial activity against pathogens.
These findings emerged in this preliminary research and highlight the probiotic potential of LAB isolated from Amazonian cocoa fermenters.
Author Contributions: Conceptualization, Laura Scalvenzi, and Roldan Torres Gutiérrez; methodology, Roldan Torres Gutiérrez, Liliana Cerda Mejía, Andrea Riofrio Carrión; resources, Laura Scalvenzi and Roldan Torres Gutiérrez; data curation, Laura Scalvenzi, Roldan Torres Gutiérrez and Manuel Pérez Quintana; writing—original draft preparation, Laura Scalvenzi and Roldan Torres Gutiérrez; writing—review and editing, Manuel Pérez Quintana, Liliana Cerda Mejía; project administration, Laura Scalvenzi; funding acquisition, Laura Scalvenzi. All authors have read and agreed to the published version of the manuscript.
Funding: Nutrition Act Co. Ltd. funded the acquisition of laboratory supplies to carry out the research.
Institutional Review Board Statement: Not applicable
Informed Consent Statement: Not applicable
Data Availability Statement: Not applicable
Acknowledgments: The manuscript's authors thank the company Nutrition Act Co. Ltd., in the persons of Ken Yamauchi and Ulrich Zebisch, and the Universidad Estatal Amazónica for the facilities provided in the research development.
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
1. Aparna Sudhakaran, V., Panwar, H., Chauhan, R., Duary, R.K., Rathore, R.K., Batish, V.K., Grover, S. Modulation of anti-inflammatory response in lipopolysaccharide stimulated human THP-1 cell line and mouse model at gene expression level with indigenous putative probiotic lactobacilli. Genes Nutr 2013, 8(6), 637-648. DOI: 10.1007/s12263-013-0347-5
2. Alard, J., Lehrter, V., Rhimi, M., Mangin, I., Peucelle, V., Abraham, A.L., Mariadassou, M., Maguin, E., Waligora-Dupriet, A.J., Pot, B., Wolowczuk, I., Grangette, C. Beneficial metabolic effects of selected probiotics on diet-induced obesity and insulin resistance in mice are associated with improvement of dysbiotic gut microbiota. Environ Microbiol 2016, 18(5), 1484–1497. DOI: 10.1111/1462-2920.13181
3. Hooper, L.V., Littman, D.R., Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336(6086), 1268–1273. DOI: 10.1126/science.1223490
4. Kaji, R., Kiyoshima-Shibata, J., Nagaoka, M., Nanno, M., Shida, K. Bacterial teichoic acids reverse predominant IL-12 production induced by certain Lactobacillus strains into predominant IL-10 production via TLR2-dependent ERK activation in macrophages. J Immunol 2010, 184, 3505-3513. DOI: 10.4049/jimmunol.0901569
5. Smith, P., Garrett, W. The Gut Microbiota and Mucosal T Cells. Front. Microbiol. 2011, 2. https://doi.org/10.3389/fmicb.2011.00111
6. Duary, R.K., Bhausaheb, M.A., Batish, V.K., Grover, S. Anti-inflammatory and immunomodulatory efficacy of indigenous probiotic Lactobacillus plantarum Lp91 in colitis mouse model. Mol. Biol. Rep. 2012, 39, 4765–4775.
7. Swain, M.R., Anandharaj, M., Ray, R.C., Parveen Rani, R. Fermented Fruits and Vegetables of Asia: A Potential Source of Probiotics. Biotechnology Research International 2014, 250424. doi:10.1155/2014/250424
8. Granato, D., Branco, G.F., Nazzaro, F., Cruz, A.G., Faria, J.A.F. Functional Foods and Nondairy Probiotic Food Development: Trends, Concepts, and Products. Compr. Rev. Food Sci. Food Saf. 2010, 9, 292–302. doi: 10.1155/2014/250424
9. Saito, V.S.T., dos Santos, T.F., Vinderola, C.G., Romano, C., Nicoli, J.R., Araújo, L.S., Costa, M.M., Andrioli, J.L., Uetanabaro, A.P.T. Viability and resistance of lactobacilli isolated from cocoa fermentation to simulated gastrointestinal digestive steps in soy yogurt. J Food Sci 2014, 79(2), 208-213. DOI: 10.1111/1750-3841.12326
10. Qamsari, E.M., Kermanshahi, R.K., Erfan, M., Ghadam, P., Sardari, S., Eslami, N. Characteristics of surface layer proteins from two new and native strains of Lactobacillus brevis. Int J Biol Macromol. 2017, 95, 1004-1010. https://doi.org/10.1016/j.ijbiomac.2016.10.089
11. Aziz, G., Fakhar, H., Rahman, S.U., Tariq, M., Zaidi, A. An assessment of the aggregation and probiotic characteristics of Lactobacillus species isolated from native (desi) chicken gut. J Appl Poultry Res 2019, 28, 846-57.
12. Ordoñez-Araque, R.H., Landines-Vera, E.F., Urresto-Villegas, J.C., Caicedo-Jaramillo, C.F. Microorganisms during cocoa fermentation: Systematic review. Foods and Raw Materials, 2020, 8.1: 155-162. http://doi.org/10.21603/2308-4057-2020-1-155-162
13. Jadan, O., Cifuentes, M., Torres, B., Selesi, D., Veintimilla, D., Gunter, S. Influence of tree cover on diversity, carbon sequestration and productivity of cocoa systems in the Ecuadorian Amazon. Bois For Trop 2015, 325(3), 35–47. DOI:10.19182/bft2015.325.a31271
14. Crafack, M., Keul, H., Eskildsen, C.E., Petersen, M.A., Saerens, S., Blennow, A.., Skovmand-Larsen, M., Swiegers, J.H., Petersen, G.B., Heimdal, H., Nielsen, D.S. Impact of starter cultures and fermentation techniques on the volatile aroma and sensory profile of chocolate. Food Res Int 2014, 63, 306–316. https://doi.org/10.1016/j.foodres.2014.04.032
15. Kongor, J.E., Hinneh, M., Van de Walle, D., Afoakwa, E.O., Boeckx, P., Dewettinck, K. Factors influencing quality variation in cocoa (Theobroma cacao) bean flavour profile - A review. Food Res Int 2016, 82, 44–52. https://doi.org/10.1016/j.foodres.2016.01.012
16. Santos, T.T., Santos, Ornellas, R.M., Borges Arcucio, L., Messias Oliveira, M., Nicoli, R., Villela Dias, C., Trovatti Uetanabaro, A.P., Vinderola, G. Characterization of lactobacilli strains derived from cocoa fermentation in the south of Bahia for the development of probiotic cultures. LWT - Food Sci Technol 2016, 73, 259–266. https://doi.org/10.1016/j.lwt.2016.06.003
17. Visintin, S., Alessandria, V., Valente, A., Dolci, P., Cocolin, L. Molecular identification and physiological characterization of yeasts, lactic acid bacteria and acetic acid bacteria isolated from heap and box cocoa bean fermentations in West Africa. Int J Food Microbiol. 2016, 216, 69-78. DOI: 10.1016/j.ijfoodmicro.2015.09.004
18. Larrea, C., Kawachi, I. Does economic inequality affect child malnutrition? The case of Ecuador. Soc Sci Med 2005, 60,165-78. https://doi.org/10.1016/j.socscimed.2004.04.024
19. De Vuyst, L., Weckx, S. The cocoa bean fermentation process: from ecosystem analysis to starter culture development. J Appl Microbiol 2016, 121(1), 5-17. https://doi.org/10.1111/jam.13045
20. De Vuyst, L, Leroy, F. Functional role of yeasts, lactic acid bacteria and acetic acid bacteria in cocoa fermentation processes. FEMS Microbiology Reviews 2020, 44(4), 432-453. doi: 10.1093/femsre/fuaa014
21. Schwan, R, Wheals, A. The microbiology of cocoa fermentation and its role in chocolate quality. Critical Reviews in Food Science and Nutrition 2004, 44(4), 205-221. doi: 10.1080/10408690490464104
22. Camu, N., De Winter, T., Verbrugghe, K., Cleenwerck, I., Vandamme, P., Takrama, J.S., Vancanneyt, M., De Vuyst, L. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl Environ Microb 2007, 73(6), 1809-1824. DOI: 10.1128/AEM.02189-06
23. De Angelis, M., Gobbetti, M. Lactic Acid Bacteria. Lactobacillus spp.: General Characteristics. Reference Module in Food Science 2016. DOI:10.1016/B978-0-12-374407-4.00259-4
24. Morante-Carriel, L., Abasolo, F., Bastidas-Caldes, C., Paz, E.A., Huaquipán, R., Díaz, R., Valdes, M., Cancino, D., Sepúlveda, N., Quiñones, J. Isolation and Characterization of Lactic Acid Bacteria from Cocoa Mucilage and Meat: Exploring Their Potential as Biopreservatives for Beef. Microbiology Research, 2023, 14(3), 1150-1167. https://doi.org/10.3390/microbiolres14030077
25. Adikari, A., Priyashantha, H., Disanayaka, J., Jayatileka, D., Kodithuwakku, S., Jayatilake, J., Vidanarachchi, J. Isolation, identification and characterization of Lactobacillus species diversity from Meekiri: traditional fermented buffalo milk gels in Sri Lanka. Heliyon 2021, 7(10), https://doi.org/10.1016/j.heliyon.2021.e08136
26. Abid, S., Farid, A., Abid, R., Rehman, M., Alsanie, W.F., Alhomrani, M., Alamri, A.S., Asdaq, S.M.B., Hefft, D.I., Saqib, S., Muzammal, M, Morshedy, S.A., Alruways, M.W., Ghazanfar, S. Identification, Biochemical Characterization, and Safety Attributes of Locally Isolated Lactobacillus fermentum from Bubalus bubalis (Buffalo) Milk as a Probiotic. Microorganisms 2022, 10(5), https://doi.org/10.3390/microorganisms10050954
27. Taye, Y., Degu, T., Fesseha, H., Mathewos, M. Isolation and Identification of Lactic Acid Bacteria from Cow Milk and Milk Products. Scientific World Journal 2021, doi: 10.1155/2021/4697445
28. Genin, M., Clement, F., Fattaccioli, A., Raes, M., Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 2015, Aug 8, 15, 577. DOI: 10.1186/s12885-015-1546-9
29. Voigt, J., Textoris-Taube, K., Wöstemeyer, J. pH-Dependency of the proteolytic formation of cocoa- and nutty-specific aroma precursors. Food Chem 2018, 255, 209-215. https://doi.org/10.1016/j.foodchem.2018.02.045
30. Ho, V.T.T., Zhao, J., Fleet, G. The effect of lactic acid bacteria on cocoa bean fermentation. Int J Food Microbiol 2015, 205, 54-67. https://doi.org/10.1016/j.ijfoodmicro.2015.03.031
31. Mudoor Sooresh, M., Willing, B.P., Bourrie, B.C.T. Opportunities and Challenges of Understanding Community Assembly in Spontaneous Food Fermentation. Foods 2023, 12, 673. https://doi.org/10.3390/foods12030673
32. Oliveira, J.S., Costa, K., Acurcio, L.B., Sandes, S.H.C., Cassali, G.D., Uetanabaro, A.P.T., Costa, A.M., Nicoli, J.R., Neumann, E., Porto, A.L.F. In vitro and in vivo evaluation of two potential probiotic lactobacilli isolated from cocoa fermentation (Theobroma cacao L.). J Func Foods 2018, 47, 184-191. https://doi.org/10.1016/j.jff.2018.05.055
33. Papalexandratou, Z., Kaasik, K., Villagra Kauffmann, L., Skorstengaard, A., Bouillon, G., Espensen, J.L., Hansen, L.H., Jakobsen, R.R., Blennow, A., Krych, L., Castro-Mejía, J.L., Nielsen, D.S. Linking cocoa varietals and microbial diversity of Nicaraguan fine cocoa bean fermentations and their impact on final cocoa quality appreciation. Int J Food Microbiol 2019, 304, 106-18. https://doi.org/10.1016/j.ijfoodmicro.2019.05.012
34. Idoui, T. Probiotic properties of Lactobacillus strains isolated from gizzard of local poultry. Iranian journal of microbiology 2014, 6, 120-26.
35. Da Cruz Pedroso Miguel, M.G., de Castro Reis, L.V., Efraim, P., Santos. C., Lima, N., Freitas Schwan, R. Cocoa fermentation: Microbial identification by MALDI-TOF MS, and sensory evaluation of produced chocolate. LWT 2017, 77, 362-69. https://doi.org/10.1016/j.lwt.2016.11.076
36. Kim, H.G., Gim, M.G., Kim, J.Y., Hwang, H.J., Ham, M.S., Lee, J.M., Hartung, T., Park, J.W., Han, S.H., Chung, D.K. Lipoteichoic acid from Lactobacillus plantarum elicits both the production of interleukin-23p19 and suppression of pathogen-mediated interleukin-10 in THP-1 cells. FEMS Immunol Med Microbiol 2007, 49, 205-14. DOI: 10.1111/j.1574-695X.2006.00175.x
37. Wang, Z., Xue, K., Bai, M., Deng, Z., Gan, J., Zhou, G., Qian, H., Bao, N., Zhao, J. Probiotics protect mice from CoCrMo particles-induced osteolysis. Int J Nanomed 2017, 12, 5387-5397. DOI: 10.2147/IJN.S130485
38. Lanaud, C., Vignes, H., Utge, J., Valette, G., Rhoné, B., Garcia Caputi, M., Angarita Nieto, N.S., Fouet, O., Gaikwad, N., Zarrillo, S., Powis, T.G., Cyphers, A., Valdez, F., Olivera Nunez, S.Q., Speller, C., Blake, M., Valdez, F.Jr., Raymond, S., Rowe, S.M., Duke, G.S., Romano, F.E., Loor Solórzano, R.G., Argout, X. A revisited history of cacao domestication in pre-Columbian times revealed by archaeogenomic approaches. Scientific Reports 2024, 14(1), https://doi.org/10.1038/s41598-024-53010-6
Received: 1 August 2024/ Accepted: 1 September 2024 / Published: 15 September 2024
Citation: Scalvenz L, Torres Gutiérrez R , Cerda Mejía L , Riofrio Carrión A , Pérez Quintana M. Preliminary Assessment of the In Vitro Immunological Activity of Native Lactobacillus Strains from the Ecuadorian Amazon. Bionatura Journal 2024; 1 (3) 20. http://dx.doi.org/10.70099/BJ/2024.01.03.20
Correspondence should be addressed to lscalvenzi@uea.edu.ec
Peer review information. Bionatura thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
ISSN. 3020-7886
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura Journal stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).