Taxonomic interpretation of non-heterocystous Cyanobacteria (Oscillatoriales) from eastern India with particular emphasis on Lyngbya Plectonema complex

Sreemanti Banerjee1*, Akanksha Singh2, Ruma Pal3
1Department of Botany, Diamond Harbour Women's University, Diamond Harbour Road, Sarisha, South 24 Parganas, Pin- 743368.
2Department of Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur-721302.
3Department of Botany, University of Calcutta, 35 Ballygunge Circular Road, Kolkata-700019.
*Corresponding author- sreemantibanerjee@gmail.com
Available from. http://dx.doi.org/10.21931/BJ/2024.01.01.66
ABSTRACT
Filamentous non-heterocystous cyanobacterial taxa from 8 genera were collected from different ecological niches like high altitudes, plains and estuaries of eastern India. The systematic accounts of 23 taxa were investigated
Keywords: Cyanobacteria; ITS; Oscillatoriaceae; Phormidiaceae; Phylogenetic tree; Pseudanabaenaceae.
INTRODUCTION
The filamentous cyanobacteria without heterocyst and akinetes are generally grouped together in the order Oscillatoriales 1 . This large group has simple trachomatous members with or without the sheath. Besides different morphological traits like sheath morphology, cell shape, size and cross-wall constriction, several other characteristics were considered from time to time for proper identification of cyanobacterial taxa like nucleoid structure 2–4 , fimbriae 5 , membrane system architecture 6 , composition of cell wall external structure 7,8 , consequences of cyanophage infection 9 and polyphosphate bodies 10–12 . Monumental taxonomic works have been done to classify the group according to their morphological perspectives 13,14 . Both non-heterocystous and heterocystous genera were classified under the same order, Nostocales, where non-heterocystous filamentous cyanobacteria were grouped into the family Oscillatoriaceae with a total of 15 genera and 250 species 14 . The genera Lyngbya, Phormidium, and Oscillatoria dominated the group, having 65, 45, and 76 species, respectively. Relying solely on morphological traits, few misidentifications at the generic level were detected in classical taxonomy after the intervention of molecular methods along with ultrastructure of thylakoids and cultural behavior studies.1 Based on this, a number of genera like Lyngbya, Oscillatoria, Leptolyngbya, and Phormidium were reconsidered and split into many new genera to reduce confusion. However, the genus Oscillatoria is still understudied compared to others 15 . For cyanobacteria, 16S rDNA, 16S-23S ITS region, phycocyanin gene, if D, psbA, rbcLX, and rpoC1 are considered the significant molecular markers used for the determination of phylogenetic relationship in taxonomically complex group 16–20 . Though 16S rDNA is widely used for oscillations, the reliability of 16S rDNA has been restricted to higher taxonomic groups and has not been proven efficient at the sub-generic level because of its conserved nature.21 In that context, the intergenic spacer region between the 16S-23S region was investigated to study the variation at the species level. The heterogeneities were noticed mainly in the size, secondary structure, and, most importantly, insertion and deletion of tRNA genes in multiple operons 22 . ITS region was less
Phylogenetic relationships using intergenic spacer 23 . Boyer et al. (2001)24 compared the variations in the ITS region for the first time in five heterocystous genera. The ITS region should always be handled accurately to avoid 22 . Instead of directly constructing a phylogenetic tree, secondary structures of the ITS region should also be used for better resolution at the molecular level. Due to a few earlier trials worldwide, no such threshold similarity percentage of the ITS region is fixed for concluding conspecificity in the order Oscillatoriales.
MATERIALS AND METHODS
Sample collection
Cyanobacterial samples were collected from 23 sites in Eastern India (Figure 1). The latitude and longitude of the collection sites are represented in Table 1. The sites were located in northern hilly areas, suburb areas, lateritic soil areas, hot spring habitats,
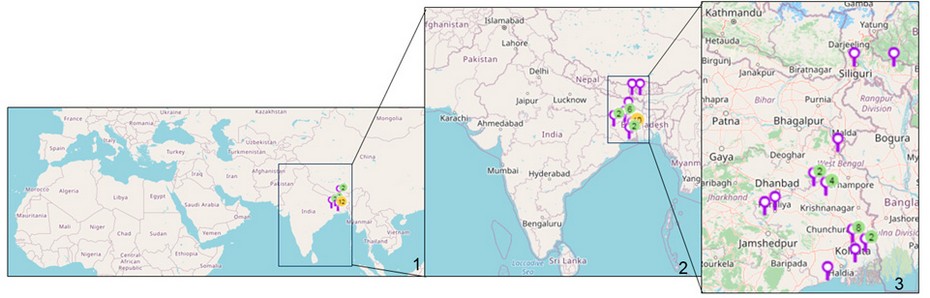
Figure 1: Map of sampling locations of Cyanobacteria in India. 1. Location sites are indicated. 2. Enlarged view of India showing the collection sites. 3. Enlarged view of eastern India showing the collection sites. (Map courtesy- Mapline, mapping software).
Immediate field vouchers were prepared at the sampling site using 4% formaldehyde in 25×50mm screw cap bottles (Tarson). The vouchers were deposited in the Calcutta University Herbarium for Algae (CUH/AL). The voucher numbers were assigned and are represented in Table 2. The remaining samples were cleaned thoroughly in double distilled water 2-3 times to remove debris from filaments. A portion of cleaned filaments was soaked in blotting paper, and approximately 100 mg of dried filaments were kept at -20ºC for molecular characterization
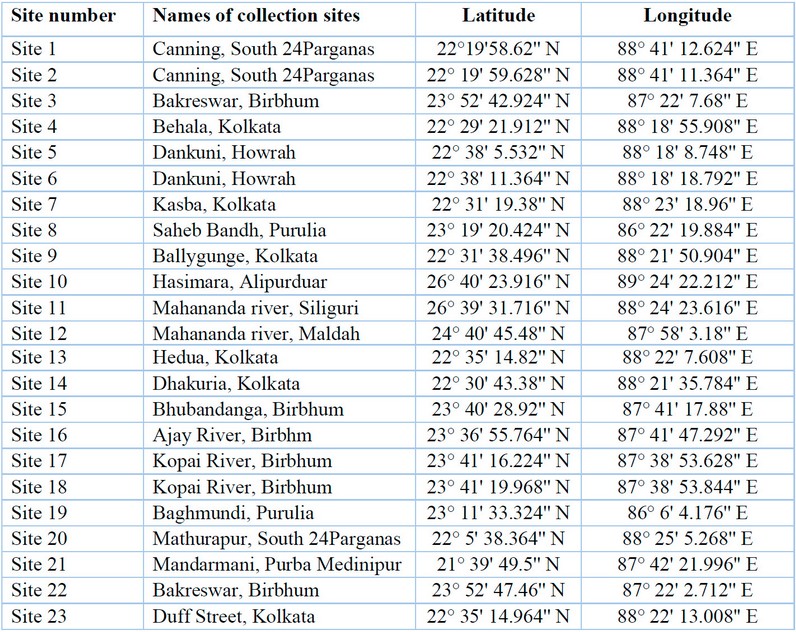
Table 1: Sample collection sites, along with their latitudes and longitudes.
Morphological characterization
The slides were prepared, and morphological details were studied using Carl Zeiss Axiostar microscope with 40X and 100X objective lenses. Photomicrographs were taken using Canon T2-T2 1, 6x SLR 426115. Morphological identifications were performed using Komarek and Anagnostidis (2005) and 25 .
Molecular characterization
The stored samples were subjected to genomic DNA extraction following phenol-chloroform extraction method 26 . Extracted DNA was stored in Tris-EDTA buffer at -20ºC. Partial 16S rDNA and 16S-23S ITS regions were amplified using suitable primers (Table 2) 27,28 . The amplifications were carried out in a thermal cycler machine (Biorad T100 PCR thermal cycler). The PCR cycle conditions were initial denaturation for 5 min at 94ºC, 35 cycles of 94ºC for 30 s, 45 s at 52ºC for ITS region and 58.5ºC for 16S rDNA region, 1 min at 72ºC, and a final extension step for 10 min at 72ºC. The PCR products were visualized using a gel documentation system in 1% agarose gel (Vilber). The PCR products were subjected to purification and Sanger sequencing through a commercial sequencing service (Syngex India).
Phylogenetic analysis
To 29 . The 30 .
The secondary structures of the 16S-23S internal transcribed spacer (ITS) were determined using Mfold version 2.3 with default parameters

Table 2: Primers used for amplification and sequencing.
Cultural behavior studies
The selected samples were thoroughly cleaned in sterile double distilled water and cultured in BG11 medium (NaNO3-1.5 g L-1; MgSO4.7H2O-0.075 g L-1; CaCl2.2H2O-0.036 g L-1; Na2CO3 0.02 g L-1; K2HPO4- 0.04 g L-1) at 20°C under cool fluorescent lights at 20–30 μmol photons m−2 s−1 with 16:8 h light: dark. All the cultures were kept for 15 days, and morphology was studied after subsequent time intervals. Slides were prepared, and photomicrographs were taken under 40X and 100X objective lenses using the Carl Zeiss Axiostar microscope attached with Canon T2-T2 1, 6x SLR 426115.
RESULTS
Twenty-three samples were identified based on morphological characters and grouped under 8 genera, namely Leptolyngbya, Planktothrix, Oscillatoria, Lyngbya, Phormidium, Limnothrix, Plectonema, and Desertifilum. Among them, Lyngbya was found in rivers of the northern uphill and lateritic soil regions and the Gangetic plain associated with the coastal region. Genus Oscillatoria was collected from north hilly aquatic habitats and the areas surrounding populated cities, whereas Plectonema was recorded from the aquatic habitats of lateritic soil areas. Leptolyngbya was found in the hot spring habitat and brackish mangrove region, whereas Limnothrix was detected only from the hot spring. Three genera like, Planktothrix, Phormidium, and Desertifilum, were solely found in suburban areas (Table 3). Photomicrographs of collected samples are represented in Figure 4-16 and 17-28. A total of 11 species were identified namely Leptolyngbya valderiana (Gomont) Anagnostidis et Komarek, Limnothrix vacuolifera (Skuja) Komárek ex G.McGregor, Planktothrix pseudagardhii Suda, Phormidium autumnale [Agardh] Trevisan ex Gomont, Phormidium formosum (Bory ex Gomont) Anagnostidis and Komárek, Lyngbya birgei G.M. Smith, Lyngbya aestuarii Liebman ex Gomont, Lyngbya semiplena J.Agardh ex Gomont, Plectonema tomasinianum Bornet ex Gomont, Oscillatoria princeps Vaucher ex Gomont and Oscillatoria sancta Kutzing ex Gomont. Taxonomic descriptions are provided in supplementary material.
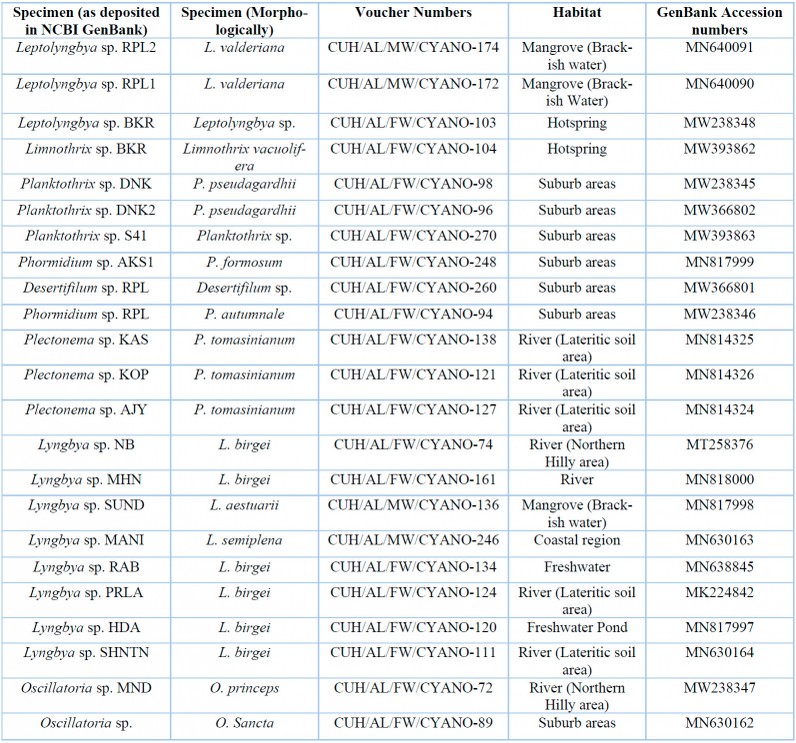
Table 3: List of specimens with their voucher numbers and GenBank accession numbers of 16S-23S ITS sequences.
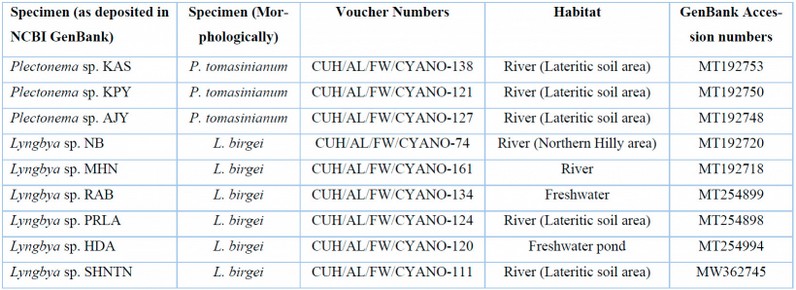
Table 4: List of freshwater Lyngbya and Plectonema samples with their voucher numbers and GenBank accession numbers of 16S sequences.
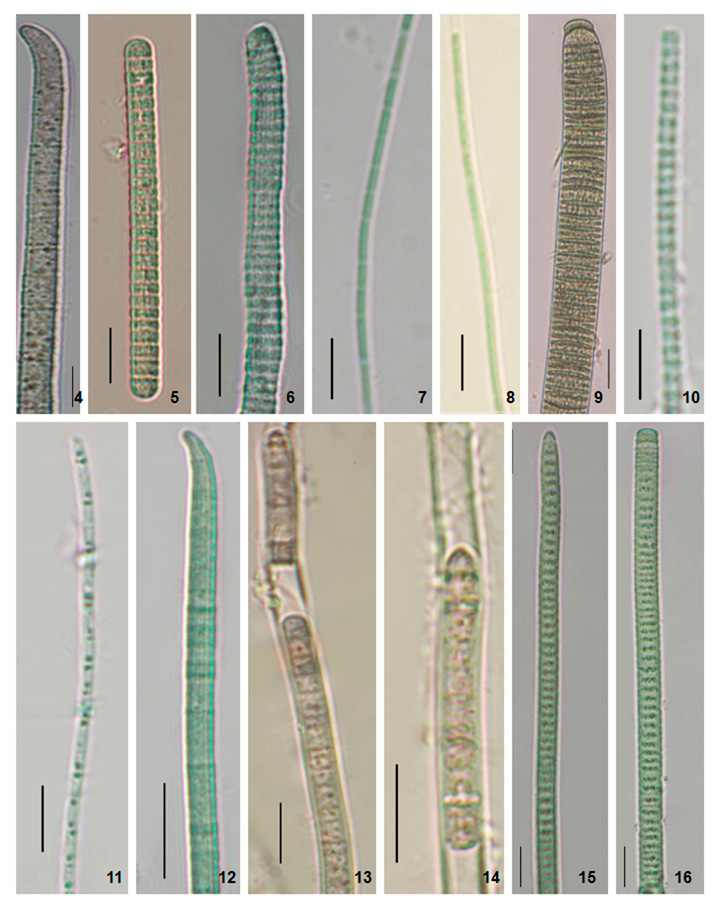
Figure 2: Photomicrographs of the specimens. 4. Phormidium formosum (Phormidium sp. AKS1). 5. Planktothrix pseudagardhii (Planktothrix sp. DNK). 6. Planktothrix pseudagardhii (Planktothrix sp. DNK2). 7. Leptolyngbya valderiana (Leptolyngbya sp. RPL1). 8. Leptolyngbya valderiana (Leptolyngbya sp. RPL2). 9. Oscillatoria sancta (Oscillatoria sp.). 10. Leptolyngbya sp. (Leptolyngbya sp. BKR). 11. Limnothrix vacuolifera (Limnothrix sp. BKR). 12. Desertifilum sp. (Desertifilum sp. RPL) 13-14. Phormidium autumnale (Phormidium sp. RPL). 15-16. Planktothrix sp. (Planktothrix sp. S41). Scale Bar- 5 µm.

Figure 3: Photomicrographs of the specimens. 17. Lyngbya birgei (Lyngbya sp. PRLA). 18. Lyngbya birgei (Lyngbya sp. NB). 19. Lyngbya birgei (Lyngbya sp. SHNTN). 20. Lyngbya birgei (Lyngbya sp. RAB). 21. Lyngbya birgei (Lyngbya sp. MHN). 22. Lyngbya birgei (Lyngbya sp. HDA). 23. Plectonema tomasinianum (Plectonema sp. KOP). 24. Plectonema tomasinianum (Plectonema sp. AJY). 25. Oscillatoria princeps (Oscillatoria sp. MND). 26. Lyngbya aestuarii (Lyngbya sp. SUND). 27. Plectonema tomasinianum (Plectonema sp. KAS). 28. Lyngbya semiplena (Lyngbya sp. MANI). Scale Bar- 5 µm.
A phylogenetic study using 16S-23S ITS region
Based on morphology, the genera were grouped into Pseudanabaenaceae, Oscillatoriaceae, and Phormidiaceae. The phylogenetic trees were constructed within each family. The ITS length was around 250bp except for the MW238347 Oscillatoria sp. MND (morphologically identified as O. princeps) was almost 450bp. The similarity matrices of the sequences are represented in Supplementary Tables 1-6.
The Pseudanabaenaceae tree was constructed from a total of 34 sequences of ITS, including 4 from our study and 30 from the NCBI database (Fig. 29). The samples were resolved into 3 distinct clades with moderate to high bootstrap support (NJ/ML) values. The sequence similarities were represented in supplementary table 1. In clade 1 brackish water taxa MN640090 Leptolyngbya sp. RPL1(morphologically L. valderiana) and MN640091 Leptolyngbya sp. RPL2 (morphologically L. valderiana) formed a monophyletic clade with high bootstrap support with other saline Leptolyngbya sequences of the NCBI database (HQ891920, MF084983, MF084982, MF084981) and showed polyphyly with the experimental MW2383488 Leptolyngbya sp. BKR (morphologically Leptolyngbya sp.) from Hotspring. The MW2383488 Leptolyngbya sp. BKR formed monophyly with a hot-spring KM438181 Leptolyngbya sp. of Greece (clade 2). The sequences of Albertania, which was recently established as a new genus from Leptolyngbya, showed similarity with the Leptolyngbya sp. BKR and shared sister clade with the experimental taxa. Our experimental MW393862 Limnothrix sp. BKR (morphologically Limnothrix vacuolifera) sequence was well separated from the Leptolyngbya clades and formed a monophyletic clade with Limnothrix, Geitlerinema, and Jaaginema having high bootstrap values (clade 3).
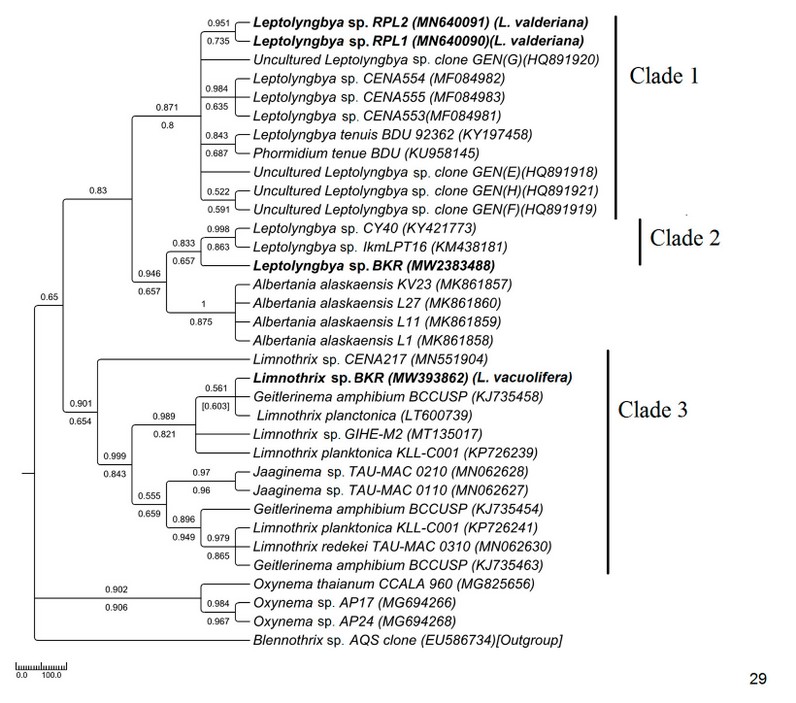
Figure 4: Phylogenetic tree of ITS sequences based on neighbor-joining and the maximum likelihood of the family Pseudanabaenaceae. Bootstrap values (NJ/ML) for nodes >50% were shown in the tree. Genbank accession numbers are provided.
The phylogenetic tree of Phormidiaceae was built up with 29 NCBI sequences along with 6 sequences of the current study (Fig. 30).
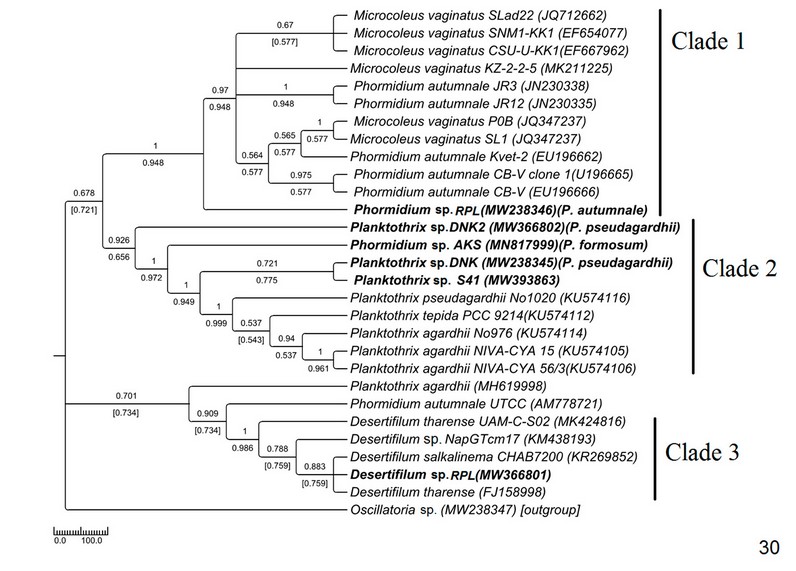
Figure 5: Phylogenetic tree of ITS sequences based on neighbor-joining and the maximum likelihood of the family Phormidiaceae. Bootstrap values (NJ/ML) for nodes >50% were shown in the tree. Genbank accession numbers are provided.
Phylogenetic relationships in Oscillatoriaceae
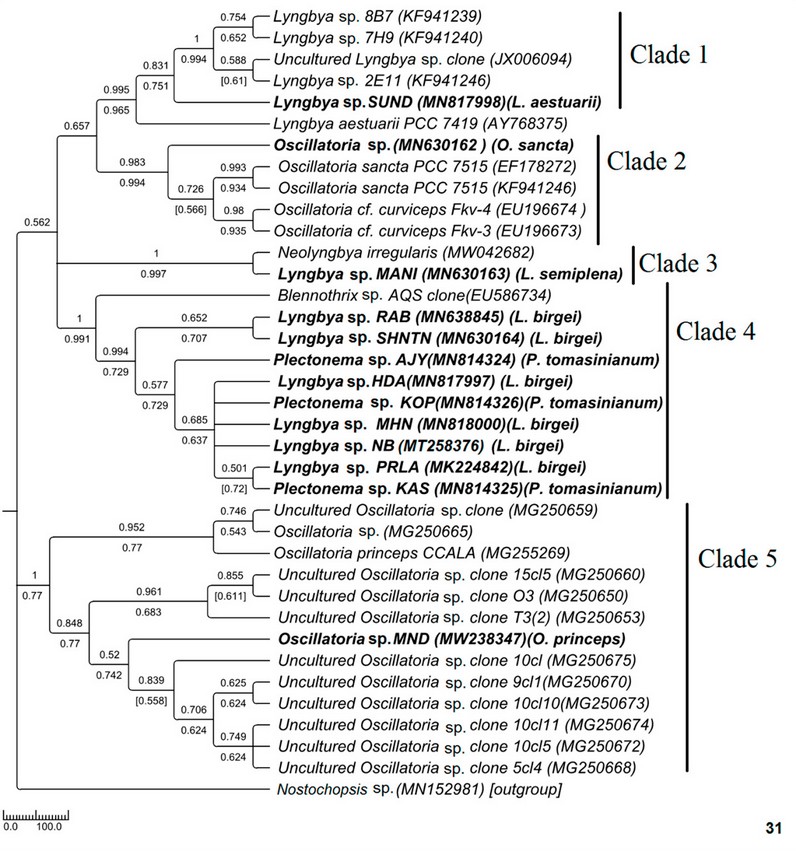
Figure 6: Phylogenetic tree of ITS sequences based on neighbor-joining and the maximum likelihood of the family Oscillatoriaceae. Bootstrap values (NJ/ML) for nodes >50% were shown in the tree. Genbank accession numbers are provided.
Phylogenetic study in Oscillatoriaceae using 16S rDNA region
The average sequence length in the tree was around 700bp. Samples
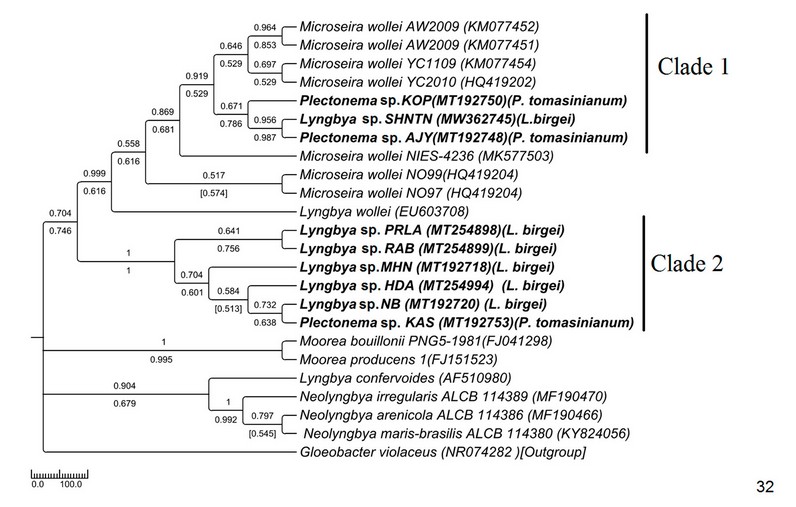
Figure 7: Phylogenetic tree of 16S sequences based on neighbor-joining and the maximum likelihood of the family Oscillatoriaceae. Bootstrap values (NJ/ML) for nodes >50% are shown in the tree. Genbank accession numbers are provided.
Cultural behavior study of Lyngbya-Plectonema complex
The cultural behavior of Plectonema tomasinianum samples and Lyngbya birgei were studied for further clarification. Surprising similarities in different growth stages were recorded among them. On the same note, Lyngbya samples formed false branching like Plectonema samples during the growth of filaments. The detailed changes are in Table 5, Figures 33-42, and Figures 43-57.
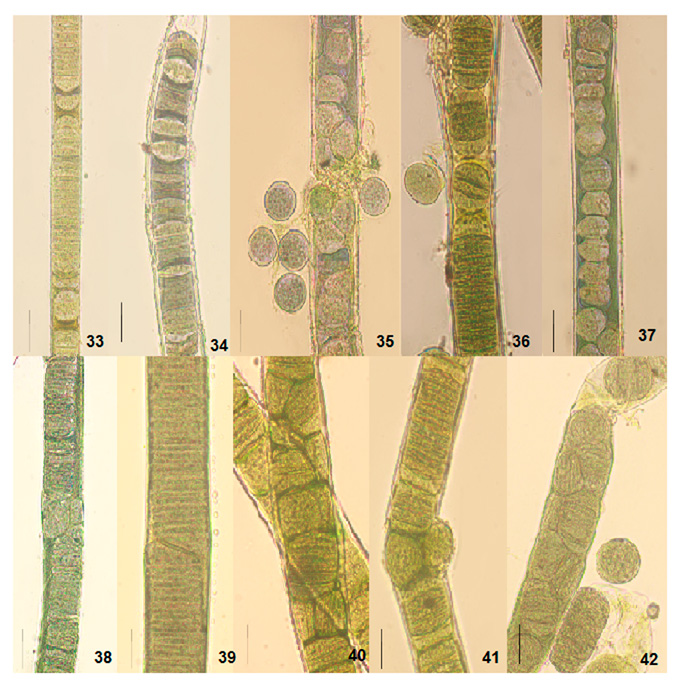
Figure 8: Light photomicrographs of Lyngbya birgei and Plectonema tomasinianum filaments at different growth phases in culture show the formation of separation discs and pseudohormogone formation. 33. Separation disc formation in Lyngbya sp. RAB filament. 34. Separation disc formation in Lyngbya sp. HDA filament. 35. Pseudohormogone formation in Plectonema sp. KAS. 36. Hormogone and pseudohormogone formation in Plectonema sp. KPY. 37. Pseudohormogone formation in Plectonema sp. AJY. 38. Cell division in different planes in Lyngbya sp. RAB. 39. Cell division in other planes in Lyngbya sp. HDA. 40. Cell division in different planes in Plectonema sp. KAS. 41. Cell division in different planes in Plectonema sp. KPY. 42. Cell division in other planes in Plectonema sp. AJY. Scale bar- 20 µm.
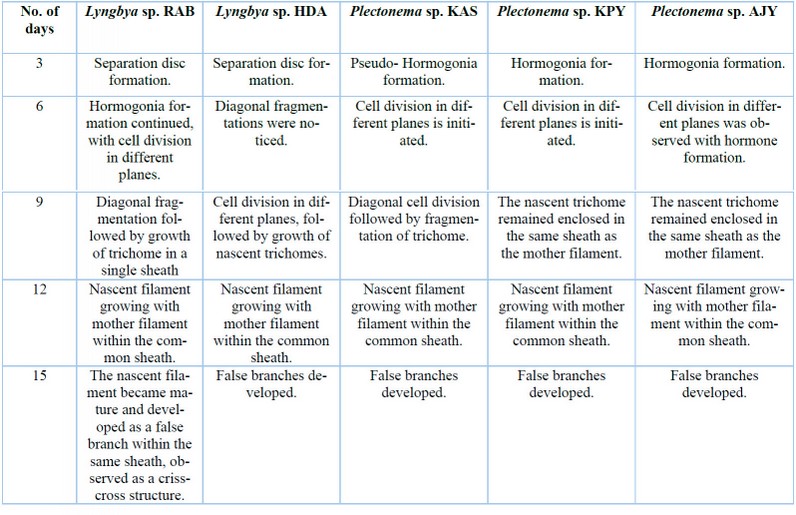
Table 5: Variation in growth performances of Lyngbya and Plectonema in cultural conditions.
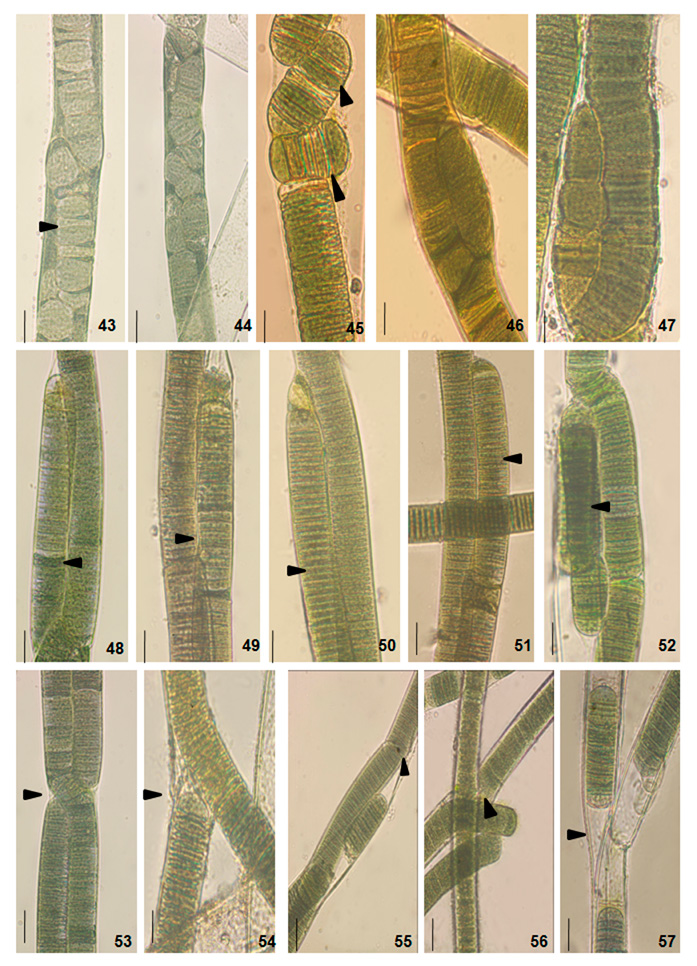
Figure 9: Light photomicrographs of Lyngbya birgei and Plectonema tomasinianum filaments at different growth phases in culture show diagonal fragmentation followed by nascent trichomes and false branch development initiation. Arrows indicate the changes. 43. Filament of Lyngbya sp. RAB. 44. Filament of Lyngbya sp. HDA. 45. Filament of Plectonema sp. KAS. 46. Filament of Plectonema sp. KPY. 47. Filament of Plectonema sp. AJY. 48. Filament of Lyngbya sp. RAB. 49. Filament of Lyngbya sp. HDA. 50. Filament of Plectonema sp. KAS. 51. Filament of Plectoneme sp. KPY. 52. Filament of Plectonema sp. AJY. 53. Filament of Lyngbya sp. RAB. 54. Filament of Lyngbya sp. HDA. 55. Filament of Plectoneme sp. KAS. 56. Filament of Plectonema sp. KPY. 57. Filament of Plectonema sp. AJY. Scale bar- 20 µm.
ITS secondary structure analysis
The 16S-23S ITS regions were subjected to the identification of conserved
DISCUSSION
The current approaches to cyanobacterial systematics faced considerable challenges in building up conclusive 24 . In the present study, 23 taxa under 8 genera of the order Oscillatoriales were investigated
To date, genera of non-heterocystous Oscillatoriales have yet to be well resolved by molecular approach. The Leptolyngbya is one of the most abundant and complicated genera of Pseudanabaenaceae due to its simple morphology with genetic variation1 . A few Lyngbya, Plectonema and Phormidium members with very thin trichomes (0.5-3.5µm wide) were described as a new genus Leptolyngbya33 . The monophyletic assemblage of this genus has not been proved yet. Polyphyletic assemblage based on 16S rDNA analysis was observed in the case of Pseudanabaena, Leptolyngbya, Limnothrix, and Geitlerinema 34,35 . In our present study, the polyphyletic origin of Leptolyngbya and Limnothrix, as well as habitat-specific clustering among species of Leptolyngbya, was noticed (Fig. 29). The sequences were clustered with other sequences with similar types of ecological preferences. Cyanobacterial habitat selectivity is a matter of debate to many researchers. The argument regarding endemism 36 or cosmopolitan distribution has existed 37 . However, present-day research has demonstrated that habitat specificity or nutrient requirement is related to genome size 38,39 . In the current study, genetic diversity among cyanobacteria dwelling in freshwater as well as extreme environments, like marine and hot springs, was studied based on ITS-based phylogenetic analysis for the very first time, which indicated that this molecular maker is capable of discriminating species belonging to different habitats.
Morphologically ambiguous identification is inevitable in the family Phormidiaceae of Oscillatoriales due to its misleading nature of identifying criteria. The name of Phormidium was considered an invalid name due to more than 100 years of taxonomic confusion40 . However, Casamatta et al. (2005)41 did not agree with the fact totally but concluded that generic and subgeneric categorizations in this genus should not essentially correspond to systematic relations. The genus was revised and grouped using morphological and ultrastructural criteria to ease the identification process1 . The 16S rDNA-based analysis showed a phylogenetically close Microcoleus vaginatus and Phormidium autumnale assemblage with almost indistinguishable morphological characters34 . Both the taxa mainly differed in their sheath properties and filament size influenced by environmental factors; therefore, they are inconsistent in culture conditions and 16S rDNA-based phylogenetic study33 . In the current ITS sequence-based study also, the Microcoleus and Phormidium formed a monophyletic assemblage that appeared as sister to the experimental genus Phormidium (morphologically P. autumnal), indicating congruency between 16S and ITS sequence-based interpretations (Fig. 30).
The genus Planktothrix was placed under Phormidiacae (after separation from Oscillatoria) because of their cellular characteristics and support from 16S rDNA sequencing42 . In our study, the placement of MW238346 Phormidium sp. (morphologically P. formosum) with Planktothrix sequences (Fig. 30) justified polyphyly of Phormidium as proved by other authors also42 . This kind of polyphyly may have resulted due to inadequate deposited sequences, and thus, more ITS sequence-based research should be done to enrich the sequence database. Paraphyly of Planktothrix was observed in our study (Fig. 30) as interpreted in earlier studies using 16S rDNA42 . Desertifilum was considered a new genus under Phormidiaceae after being separated from Microcoleus and Phormidium based on 16S rDNA-based analysis coupled with morphological parameters43 . In the current study, Desertifilum was in a monophyletic lineage consistent with their morphology and separated from the Microcoleus-Phormidium clade. The interpretation is congruent with the 16S rDNA-based study by the earlier workers. This is the first report of Desertifilum from eastern India.
Polyphyletic nature of Oscillatoria was also reflected in this ITS sequence-based study (Fig. 31). In the phylogenetic tree of Oscillatoriaceae, MN630162 Oscillatoria sp. (morphologically O. sancta) having apical cap was placed in monophyletic lineage with other O. sancta sequences with high sequence similarity and good bootstrap support whereas MW238347 Oscillatoria sp. MND (morphologically O. princeps) formed a well-supported clade with other Oscillatoria strains and was distantly placed from O. Sancta. As studied by Hasler et al. (2012), 34 Oscillatoria sancta having thickened apical cap formed well-supported lineage with other calyptrate taxa in 16S rDNA-based study. The origin of calyptra or their role in cyanobacteria has never been investigated44 . Phylogenetic remoteness, as observed in our study, cannot be disregarded as both the sequences have shared a high level of similarity with their sister sequences having good bootstrap support. The genus Oscillatoria was not revised earlier; however, from our observation, it can be concluded that Oscillatoria having an apical cap showed a somewhat different evolutionary history from that of Oscillatoria without an apical cap. Further detailed investigation is needed in this regard.
For Lyngbya, brackish water and marine species were well separated, and their monophyletic assemblage with other ecologically consistent strains supported again that ITS-based sequence clustering can reflect ecological selectivity. Genetic divergence can reflect habitat preferences for ecologically distinct organisms45 . In the phylogenetic tree of Oscillatoriaceae, freshwater Lyngbya sp. (morphologically L. birgei) formed a different monophyletic lineage than that of marine Lyngbya (Fig. 31). The morphological similarity was also tested at the molecular level because all the samples have shown 99-100% similarity. Surprisingly, Plectonema shared a common monophyletic lineage with the Lyngbya clade, having 99-100% sequence similarity. Though no threshold percentage value is available in the literature for species-level identification, the high-level sequence similarity of the ITS study may be considered further.
The ITS tree topology was compared with rDNA based phylogenetic tree (Fig. 32), and the topology was almost similar. However, two Plectonema samples (MT192748 Plectonema sp. AJY and MT192750 Plectonema sp. KPY) (morphologically P. tomasinianum) and MW362745 Lyngbya sp. SHNTN, all collected from rivers flowing over lateritic soil regions, formed a monophyletic clade with Microseira wollei, a nascent genus separated from Lyngbya. MT192753 Plectonema sp. KAS collected from similar habitats and having exactly similar morphology have been placed in the Lyngbya clade with 98% similar sequences. As per convention, more than 98% 16SrDNA similarity can be considered the same species46 . Thus, in our present study, morphological identification of freshwater Lyngbya sequences from high to low altitudes became congruent with molecular characterization based on ITS and 16S sequence data. Both morphological and molecular details support that all freshwater Lyngbya belong to the same species. However, a problem in identifying Plectonema still needs to be resolved. Secondary structures (Table 6, Supplementary Figure 1) were also investigated, but no differences were noticed among the samples.
Plectonema was initially classified under Scytonemataceae14,47 based on false branching formation, though no heterocyst was found. Genus Lyngbya was classified under Oscillatoriaceae by Geitler (1932), where the occasional occurrence of false branching was also noticed. Later on, the observation of other scientists inferred that false branching occurred facultatively in a tiny part of the population of P. tomasinianum, and the character was also overcredited, which should not be done because of its inconsistency48–50 . Scarcity in the P. tomasinianum sequence in online databases led to the dilemma that existed for a long. In the present study, false branching in Lyngbya culture again confounded the generic discrimination between Plectonema tomasinianum and Lyngya birgei. Supporting the earlier workers, the current study also emphasized and proposed that this kind of plastic, pleomorphic character of cyanobacteria should not be considered as an autapomorphic character at the genus level.
CONCLUSIONS
The study marks the first-ever attempt to use ITS-based phylogenetic analysis on Indian non-heterocystous filamentous cyanobacteria. The results of our research demonstrate the impressive discriminatory ability of the 16S-23S ITS region in identifying and distinguishing cyanobacteria from various habitats. Our study highlights the novelty of this approach and its potential for revolutionizing the field of cyanobacterial research. Our research shows that species clustering in the ITS-based phylogenetic tree is habitat-specific. For instance, marine species of Leptolyngbya form a monophyletic clade with other marine Leptolyngbya sp., while hotspring taxa form a separate monophyletic clade with other hotspring Leptolyngbya species. Moreover, the study revealed the polyphyletic nature of Planktothrix and Phormidium in the phylogenetic tree of Phormidiaceae. The ITS-based species clustering supported earlier 16S rDNA-based studies, highlighting the significance of the ITS region in species identification.
We present a novel approach to differentiate various species of Lyngbya by proposing 98% sequence similarity as a threshold percentage of ITS sequence. Furthermore, this study emphasizes the need to re-evaluate the taxonomic status of Lyngbya birgei and Plectonema tomasinianum as they appear highly similar based on 16S rDNA and ITS sequence-based analysis and cultural behavior study. Our study offers valuable insights into the complexities of identifying these species and calls for more research in this area.
Figure S1: Secondary structures of 16S–23S internal transcribed spacer (ITS) sequences from the Lyngbya sp.HDA, Lyngbya sp.RAB, Plectonema sp. KOP, Plectonema sp. KAS and Plectonema sp. AJY., Table S1: A partial similarity matrix (P distance) generated using 16S-23S ITS region (Family- Pseudanabaenaceae), Table S2-A partial similarity matrix (P distance) generated using 16S-23S ITS region (Family- Phormidiaceae), Table S3-A partial similarity matrix (P distance) generated using 16S-23S ITS region (Family- Oscillatoriaceae-genus Oscillatoria), Table S4-A partial similarity matrix (P distance) generated using 16S-23S ITS region (Family- Oscillatoriaceae-genus Lyngbya(marine)), Table S5-A partial similarity matrix (P distance) generated using 16S-23S ITS region (Family- Oscillatoriaceae-genus Lyngbya-Plectonema(freshwater)), Table S6-A partial similarity matrix (P distance) generated using 16S region (Family Oscillatoriaceae-genus Lyngbya-Plectonema)
Author Contributions: "Conceptualization, Sreemanti Banerjee and Ruma Pal.; methodology, Sreemanti Banerjee.; software, Sreemanti Banerjee; validation, Sreemanti Banerjee, Akanksha Singh and Ruma Pal; formal analysis, Sreemanti Banerjee; investigation, Sreemanti Banerjee, Akanksha Singh; resources, Sreemanti Banerjee; data curation, Sreemanti Banerjee; writing—original draft preparation, Sreemanti Banerjee; writing—review and editing, Ruma Pal; visualization, Sreemanti Banerjee; supervision, Ruma Pal; project administration, Sreemanti Banerjee; funding acquisition, Sreemanti Banerjee and Ruma Pal. All authors have read and agreed to the published version of the manuscript."
Funding: The present research has been funded by the Council for Science and Industrial Research (CSIR) -sanction no.-09/028(0964)2015-EMR1. CAS Phase VII and DST-FIST Level II, Department of Botany, University of Calcutta, supported the infrastructural facility.
Acknowledgments: The authors thank Prof. Sagarmoy Ghosh, Department of Microbiology, University of Calcutta, and Dr. Suparna Ghosh, Department of Microbiology, University of Calcutta, for their cooperation during the work.
Conflicts of Interest: The authors declare no conflict of interest. The funders had no role in the study's design, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
REFERENCES
1. Komarek J, Anagnostidis K. Cyanoprokaryota. II. teil: Oscillatoriales. In Süsswasserflora von Mitteleuropa. Büdel B, Gärdner G, Krienitz L, Schagerl M, editors. Vol. 19. München: Elsevier; 2005. 1–759 p.
2. Tschermak-Woess E, Schöller A. Verteilung und Aufteilung der DNS bei einigen Cyanophyceen, festgestellt durch ihre DAPI-Fluoreszenz. Plant Systematics and Evolution. 1982;140:207–23.
3. Cepák V. Morphology Of DNA containing structures (nucleoids) as a prospective character in cyanophyte taxonomy. J Physiol. 1993 Dec 20;29(6):844–52.
4. Cepák V, Kováčik L, Komárek J. DNA containing structures (nucleoids) in some cyano-phytes/cyanobacteria as revealed by DAPI fluorescent staining. Algological Studies/Archivfür Hy-drobiologie. 1991;Supplement Volumes:25–39.
5. Guglielmi G, Cohen-Bazire G. Etude taxonomique d’un genre de cyanobacterie oscillatoriacee: le genre Pseudanabaena Lauterborn. II. Analyse de la composition moleculaire et de la structure des phycobilisomes. Protistologica. 1984;20:393–413.
6. Golecki JR, Drews G. Supramolecular organization and composition of membranes. In: Carr N.G., Whitton B.A, editors. The Biology of Cyanobacteria. Blackwell Science; 1982. p. 125–41.
7. Weckesser J, Drews G, Mayer H. Lipopolysaccharides of Photosynthetic Prokaryotes. Annu Rev Microbiol. 1979 Oct;33(1):215–39.
8. Lounatmaa K, Vaara T, Österlund K, Vaara M. Ultrastructure of the cell wall of a Synechocystis strain. Can J Microbiol. 1980 Feb 1;26(2):204–8.
9. Mamkaeva KA, AL-MusAvI RA, Gromov BV. The effect of photosynthesis on the ultrastructure of a cyanophage during its intracellular growth. Mikrobiologiia. 1980;49:631–3.
10. Jensen TE, Baxter M. Imaging cell inclusions in air dried unfixed cyanobacteria with a transmission electron microscope at 75 kV. Microbios. 1985;28:145–50.
11. Baxter M, Jensen TE. Cell volume occupied by polyphosphate bodies during the polyphosphate overplus phenomenon in Plectonema boryanum. Cytobios. 1986;45:147–60.
12. Rippka R, Deruelles J, Waterbury JG, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;1–61.
13. Gomont M. Monographie des Oscillariées. Annales des Sciences Naturelles; Botanique. 1892;16:198–246.
14. Desikachary TV. Cyanophyta. Delhi: Indian Council of Agricultural Research; 1959. 1–686 p.
15. Casamatta DA, Vis ML, Sheath RG. Cryptic species in cyanobacterial systematics: a case study of Phormidium retzii (Oscillatoriales) using RAPD molecular markers and 16S rDNA sequence data. Aquat Bot. 2003 Dec;77(4):295–309.
16. Turner S. Molecular systematics of oxygenic photosynthetic bacteria. In 1997. p. 13–52.
17. Wilmotte A. Molecular evolution and taxonomy of the cyanobacteria. In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Boston: Kluwer Academic Press; 1994. p. 1–25.
18. Singh P, Singh SS, Elster J, Mishra AK. Molecular phylogeny, population genetics, and evolution of heterocystous cyanobacteria using nifH gene sequences. Protoplasma. 2013 Jun 23;250(3):751–64.
19. Mondal SK, Shit S, Kundu S. A comparative computational study of the 'rbcL'gene in plants and in the three prokaryotic families—Archaea, cyanobacteria and proteobacteria. Indian J Biotechnol. 2013;12:58–66.
20. Singh P, Fatma A, Mishra AK. Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcl gene sequence data. Ann Microbiol. 2015 Jun 30;65(2):799–807.
21. Garcı́a-Martı́nez J, Acinas SG, Antón AI, Rodrı́guez-Valera F. Use of the 16S–23S ribosomal genes spacer region in studies of prokaryotic diversity. J Microbiol Methods. 1999 May;36(1–2):55–64.
22. Boyer SL, Johansen JR, Flechtner VR, Howard GL. Phylogeny and genetic variance in terrestrial Microcoleus (Cyanophyceae) species based on sequence analysis of the 16s rrna gene and associated 16S–23S its region. J Phycol. 2002 Dec 19;38(6):1222–35.
23. Otsuka S, Suda S, Li R, Watanabe M, Oyaizu H, Matsumoto S, et al. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol Lett. 1999 Mar;172(1):15–21.
24. Boyer SL, Flechtner VR, Johansen JR. Is the 16S–23S rRNA Internal Transcribed Spacer Region a Good Tool for Use in Molecular Systematics and Population Genetics? A Case Study in Cyanobacteria. Mol Biol Evol. 2001 Jun 1;18(6):1057–69.
25. Guiry MD, Guiry GM. AlgaeBase. Worldwide electronic publication, National University of Ireland, Galway. 2021. http://algaebase.org.
26. Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–5.
27. Valério E, Chambel L, Paulino S, Faria N, Pereira P, Tenreiro R. Molecular identification, typing and traceability of cyanobacteria from freshwater reservoirs. Microbiology (N Y). 2009 Feb 1;155(2):642–56.
28. Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997 Aug;63(8):3327–32.
29. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016 Jul 1;33(7):1870–4.
30. Stöver BC, Müller KF. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010 Dec 5;11(1):7.
31. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003 Jul 1;31(13):3406–15.
32. Iteman I, Rippka R, de Marsac NT, Herdman M. rDNA analyses of planktonic heterocystous cyanobacteria, including members of the genera Anabaenopsis and Cyanospira. TheGenBank acces-sion numbers of the 16S rDNA gene sequences reported in this paper are AY038032–AY038037. Microbiology (N Y). 2002;148:481–96.
33. Stoyanov P, Moten D, Mladenov R, Dzhambazov B, Teneva I. Phylogenetic Relationships of Some Filamentous Cyanoprokaryotic Species. Evol Bioinform. 2014 Jan 23;10:EBO.S13748.
34. Hašler P, Dvořák P, Johansen JR, Kitner M, Ondřej V, Poulíčková A. Morphological and molecular study of epipelic filamentous genera Phormidium, Microcoleus and Geitlerinema (Oscillatoriales, Cyanophyta/Cyanobacteria). Fottea. 2012 Sep 1;12(2):341–56.
35. Perkerson III RB, Perkerson EA, Casamatta DA. Phylogenetic examination of the cyanobacterial genera Geitlerinema and Limnothrix (Pseudanabaenaceae) using 16S rDNA gene sequence data. Arch Hydrobiol Suppl Algol Stud. 2010 Jul 1;134:1–16.
36. Komárek J. Diversity of cyanoprokaryotes (cyanobacteria) of King George Island, maritime Antarctica - a survey. Arch Hydrobiol Suppl bd Algol Stud. 1999 Sep 16;94:181–93.
37. Mullins TD, Britschgi TB, Krest RL, Giovannoni SJ. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995 Jan 22;40(1):148–58.
38. Prabha R, Singh DP. Cyanobacterial phylogenetic analysis based on phylogenomics approaches render evolutionary diversification and adaptation: an overview of representative orders. 3 Biotech. 2019 Mar 15;9(3):87.
39. Prabha R, Singh DP, Somvanshi P, Rai A. Functional profiling of cyanobacterial genomes and its role in ecological adaptations. Genom Data. 2016 Sep;9:89–94.
40. Castenholz RW, Rippka R, Herdman M. Form genus IX. OscillatoriaVaucher 1803 (sensu Rippka, Deruelles, Waterbury,Herdman, and Stanier 1979). . In: Boone DR, Castenholz RW, editors. Bergey's Manual of Systematic Bacteriology. New York: Springer; 2001. p. 550–3.
41. Casamatta DA, Johansen JR, Vis ML, Broadwater ST. Molecular and morphological characterization of ten polar and near‐polar strains within the Oscillatoriales (Cyanobacteria). J Phycol. 2005 Apr 17;41(2):421–38.
42. Komarek J, Komarkova J. Taxonomic review of the cyanoprokaryotic genera Planktothrix and Planktothricoides. Czech Phycology, Olomouc. 2004;4:1–18.
43. Dadheech PK, Abed RMM, Mahmoud H, Mohan MK, Krienitz L. Polyphasic characterization of cyanobacteria isolated from desert crusts, and the description of Desertifilum tharense gen. et sp. nov. (Oscillatoriales). Phycologia. 2012 May 23;51(3):260–70.
44. Whitton BA. Cyanobacterial diversity in relation to the environment. In: Algal toxins: nature, occurrence, effect and detection. Dordrecht: Springer; 2008. p. 17–43.
45. Nübel U, Garcia‐Pichel F, Clavero E, Muyzer G. Matching molecular diversity and ecophysiology of benthic cyanobacteria and diatoms in communities along a salinity gradient. Environ Microbiol. 2000 Apr 21;2(2):217–26.
46. Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014 Sep 14;12(9):635–45.
47. Geitler L. Cyanophyceae in Rabenhorst’s Kryptogamen flora. Leipzing. 1932;1–1196.
48. McGregor GB, Sendall BC. Phylogeny and toxicology of Lyngbya wollei (Cyanobacteria, Oscillatoriales) from north‐eastern Australia, with a description of Microseira gen. nov. J Phycol. 2015 Feb 11;51(1):109–19.
49. Seifert M, McGregor G, Eaglesham G, Wickramasinghe W, Shaw G. First evidence for the production of cylindrospermopsin and deoxy-cylindrospermopsin by the freshwater benthic cyanobacterium, Lyngbya wollei (Farlow ex Gomont) Speziale and Dyck. Harmful Algae. 2007 Jan;6(1):73–80.
50. Speziale BJ, Turner EG, Dyck LA. “Giant” Lyngbya. Aquatics. 1988;4–11.
Received: October 9th 2023/ Accepted: January 15th 2024 / Published:15 March 2024
Citation: Banerjee S., Singh A., Pal R. Taxonomic interpretation of non-heterocystous Cyanobacteria (Oscillatoriales) from eastern India with particular emphasis on Lyngbya Plectonema complex. Revis Bionatura 2024; 1 (1) 66. http://dx.doi.org/10.21931/BJ/2024.01.01.66
Additional information Correspondence should be addressed to sreemantibanerjee@gmail.com
Peer review information. Bionatura thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
