Relationship Polymorphism of STAT5A Gene in Performance Production of Local Goats
Bashar Adham Ahmed1, Hamza Sajid Khudair2
1Assi. Prof. - Department of Animal Production - College of Agriculture - University of Diyala – Iraq
2Master student https://orcid.scopusfeedback.com/#/start
*Correspondence: basharadham@uodiyala.edu.iq
Available from. http://dx.doi.org/10.21931/BJ/2024.01.01.57
ABSTRACT
The study was conducted on a sample of 30 local goats in Diyala governorate / Canaan district in one of the goat breeders' fields in the region for the period from 13/11/2022 to 1/5/2022 to determine the genotype of the gene STAT5A EXON-7 and Its relationship to milk production (daily milk production-DMP and total milk production-TMP) and growth traits (birth weight-BWT, weaning weight-WWT and total weight gain-TWG). The analysis showed three-point mutations (CC=18 CT=12 C47064T AA=17 AT=13 A47088T and GG=15 GA=15 G47162A). There was no significant effect of the genotypes of the three mutations on all studied traits DMP, TMP, BWT, WWT, TWG, and body dimensions of the newborn at birth and weaning, body dimensions of the newborn at birth and weaning.
Keywords: Local goats, milk production, gene STAT5A EXON-7.
INTRODUCTION
Interest in raising and improving goats to benefit from milk and meat has recently begun, and goat milk is considered distinctive and healthy, especially for people who suffer from severe allergies 1, 2. Because of the increasing population numbers around the world, the demand for animal products has increased, which prompted researchers to find ways to improve the productivity of farm animals, including goats, and this was evident in many different studies 3. use of molecular genetics techniques has recently led to the discovery of genes that affect mainly and directly in many important productive and economic traits, as well as recognizing the effect of those genes by knowing the quantitative trait loci 4. Therefore, genetic markers were relied upon. In selection programs because they are more accurate than phenotypic and biochemical markers 5. Many transcription factor binding sites in the region close to the catalytic regions (promoter) have an essential effect on the transcription process, such as Nf1, CIEBP, STAT5A, and GR 6, and the STAT5A gene plays a critical role in many physiological processes. , as it is related to multiple traits such as the viability of fetuses and milk production traits 7, and the STAT5A gene is considered a mammary gland factor (Mammary Gland Factor-MGF), so it participates in the development of the mammary gland and is key in signaling the hormone prolactin ( PRL) in addition to the activity of reproducing milk protein genes 8. This gene is a transcription factor for the milk protein gene (K-casin) due to its relationship to milk production and mammary gland traits 9.
Because of the relationship of the STAT5A gene in milk production and growth traits in goats, as well as its sizeable genetic morphology, the study highlighted:
1. Determination of the genotypes of the local goats bred in Iraq based on the frequency and ratios of the genotypes of the STAT5A EXON-7 gene under study.
2. Studying the relationship of genetic polymorphism of STAT5A EXON-7 gene to animal performance in growth traits, milk production traits and its components.
MATERIALS AND METHODS
30 local Iraqi goats were used in the experiment. Molecular genetic analyses were carried out in the Biotechnology Laboratory of the Department of Animal Production / College of Agriculture / University of Diyala for the period from 2/1/2022 to 1/3/2022 to separate the genetic material (DNA) and conduct electrophoresis Polymerase chain reaction (PCR) analysis of STAT5A EXON-7 gene. The data on milk production were collected weekly for the morning circuit, and the newborn's birth weight was taken 12 hours after birth. The weaning weight was weighed after three months of birth, and the total weight gain was calculated through the weaning weight minus the birth weight, while the body dimensions of the newborns were measured. At birth and weaning according to method 10, blood samples were drawn from the jugular vein 2.5 ml using 5 ml test tubes containing Diamine Tetra acidic acid Ethylene (EDTA). After withdrawal, the blood samples were frozen at frozen temperature (-18). ) It is used in the extraction of genetic material (DNA). Kit FAVORGEN is of Taiwanese origin.
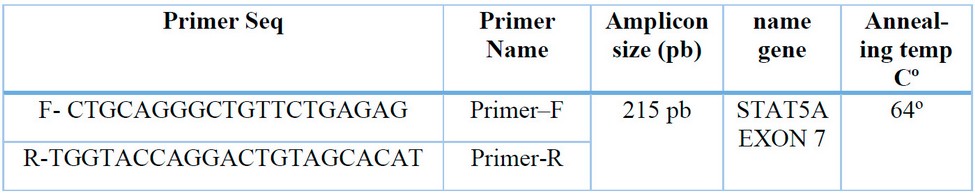
Table 1: Primer used with a length of 215 N bases of STAT5A EXON-7 gene in local goats.
After that, the studied segment of the STAT5A EXON-7 gene was detected using the polymerase chain reaction (PCR) technology. After the reaction was completed, the product of the polymerase reaction was migrated to ensure the presence of PCR products using the same method of preparing the agarose gel in the DNA migration, as DNA with a known molecular weight was loaded. (100-1500 nitrogen bases) in the first hole of the gel template and then loading the PCR product by 5 microliters into the pits of the gel template at (70V for 90 minutes) to be seen by the UV Light Transilluminator and photographing these beams. With a special camera, the packages appear colored orange ethidium bromide.
After electrophoresis, the PCR product, in a volume of 20 microliters, was sent to the Korean company Micro Gene Corporation. After obtaining the results by e-mail, Genious Software was used to analyze the results on Genebank's global website, www.ncbi.nlm.nih.go. The nucleotide sequence profile was used to determine the presence or absence of the mutation, and the curve profile was used to determine the phenotypic polymorphism of the three-point mutations of the STAT5A EXON-7 gene. The data were statistically analyzed using the statistical program SAS (Statistical Analysis System) 11 to study the relationship between the genetic structures of the STAT5A EXON-7 gene in milk production and growth traits for newborns. The test for significant differences between the means was carried out using Duncan's polynomial test 12.
Yijklm = m + Tj + Sk + Ml + eijklm
Since:
Yijklm: watch value m.
m: the general average of the adjective.
Gi = the effect of the polymorphism of the STST5A gene on the studied traits.
Tj: the effect of birth type j (single, twin).
Sk: the effect of the gender of the newborn k (male, female).
Ml: The effect of the month of birth (November, December, January, February).
Eikon: the random error that is usually and independently distributed with a mean of zero and a variance of s2e
RESULTS AND DISCUSSION
The current research detected point mutations in the studied sample of Region of STAT5A EXON-7 gene consisting of 215 bp. When analyzing the sequencing, three point mutations, C47064T A47088T and G47162A, were detected with two genotypes(CC, CT)(AA.AT) and (GG, AG), respectively.
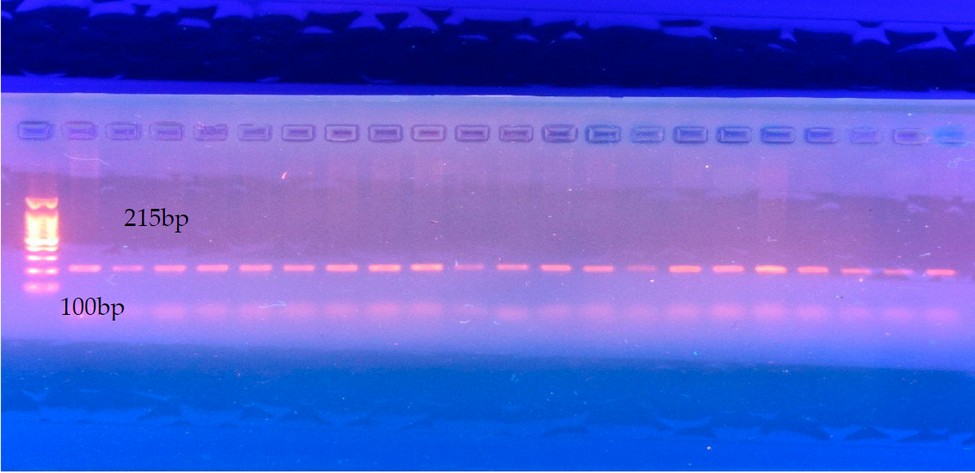
Figure 1: Shows the electrophoresis of the studied segment of the STAT5A EXON7 gene.
Were detected with two genotypes: the dominant wild CC and the hybrid CT. The frequency of the wild allele C is 0.80, While the mutant allele T is 0.20 (A47088T) with two genotypes, dominant wild AA and hybrid AT, the repeat of the wild allele A is 0.78 while the mutant allele T of 0.22, (G47162A) with two genotypes dominant wild GG and hybrid GA, the frequency of the wild allele G is 0.75 While the mutant allele C was 0.25 as shown in Table (1). A significant superiority of the chi-square value (P≤0.01) is observed for all mutations (C47064T) = 0.022 (A47088T) = 0.09 (G47162A) = 0.048.
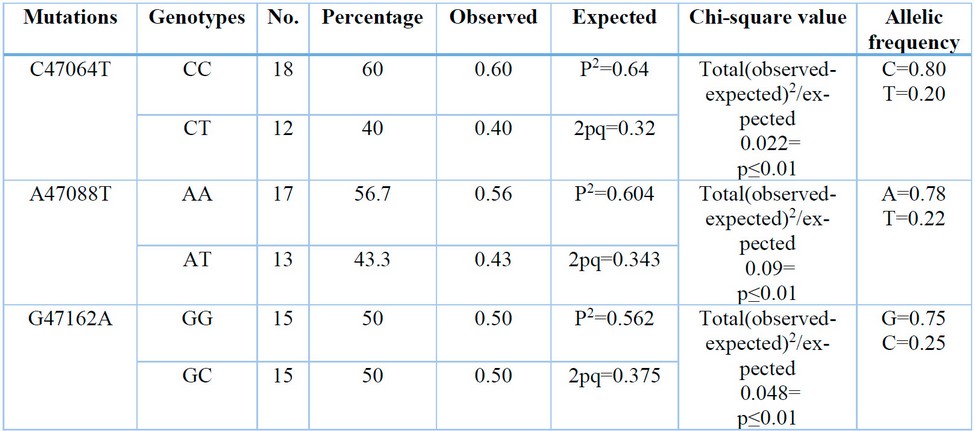
Table 2: Number, percentages of genotypes and allelic frequency of the studied mutations in the STAT5A EXON-7 gene.
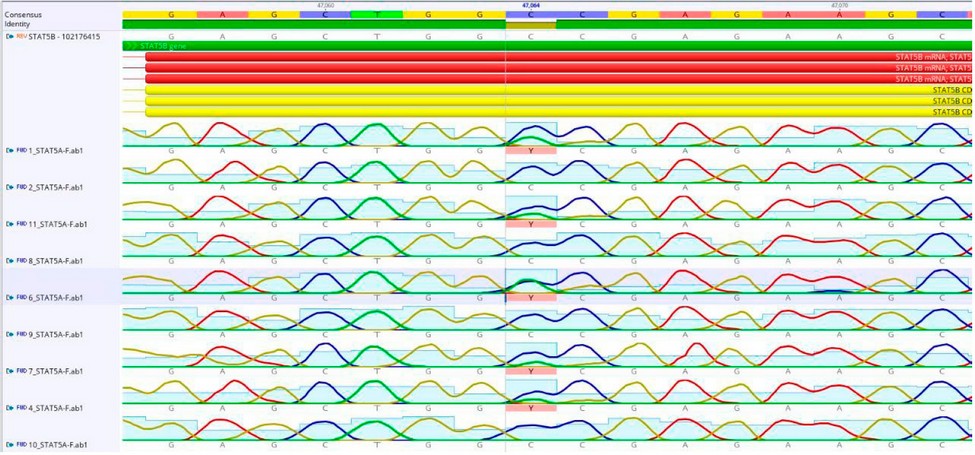
Figure 2: Shows the location of the mutation (C47064T) in the STAT5A gene EXON-7.
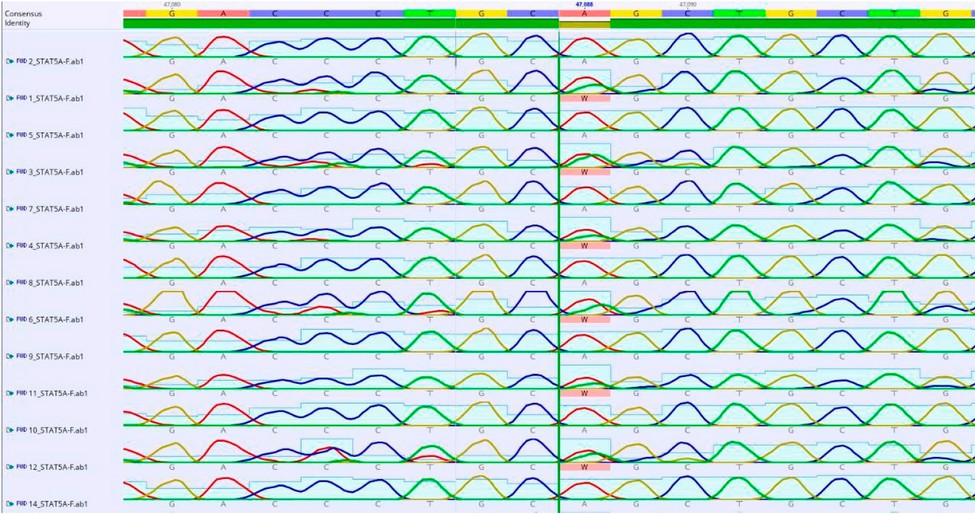
Figure 3: Shows the location of the mutation (C47088T) in the STAT5A gene EXON-7.
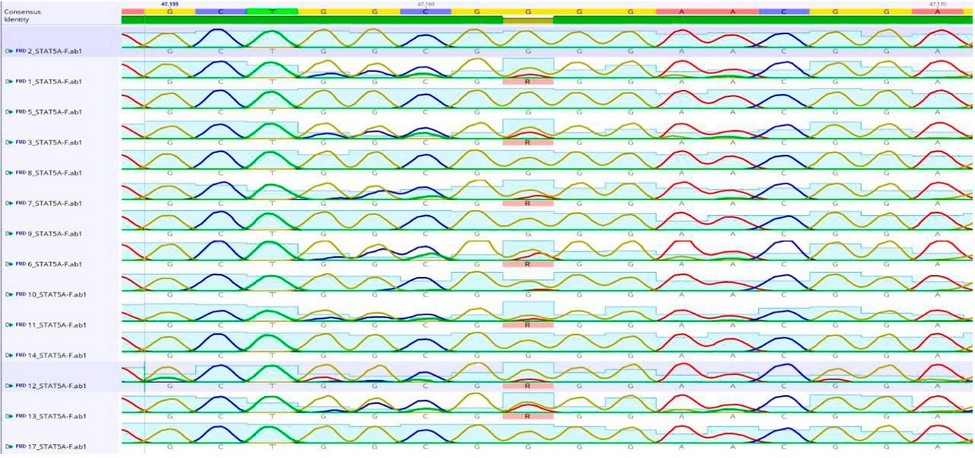
Figure 4: Shows the location of the mutation (C47062T) in the STAT5A gene EXON-7.
The results of the statistical analysis in Tables 2,3,4,5 and 6 showed that there was no significant effect of the point mutation (C47064T), (A47088T) and (G47162A) on Daily and total milk production, growth traits ( birth weight, weaning weight and total weight gain) and dimensions at birth and weaning.

Table 3: Shows the effect of genotypes in milk production for the studied point mutants.
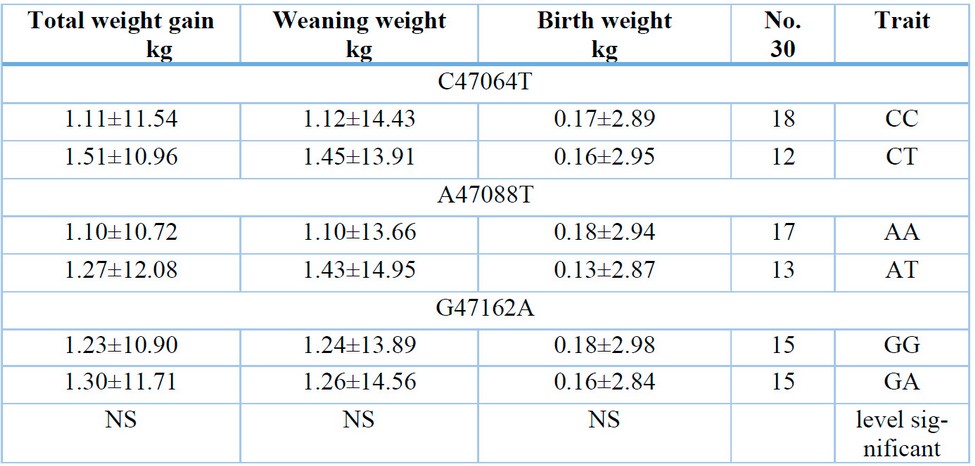
Table 4: Shows the effect of genotypes on birth weight, weaning weight and total weight gain.
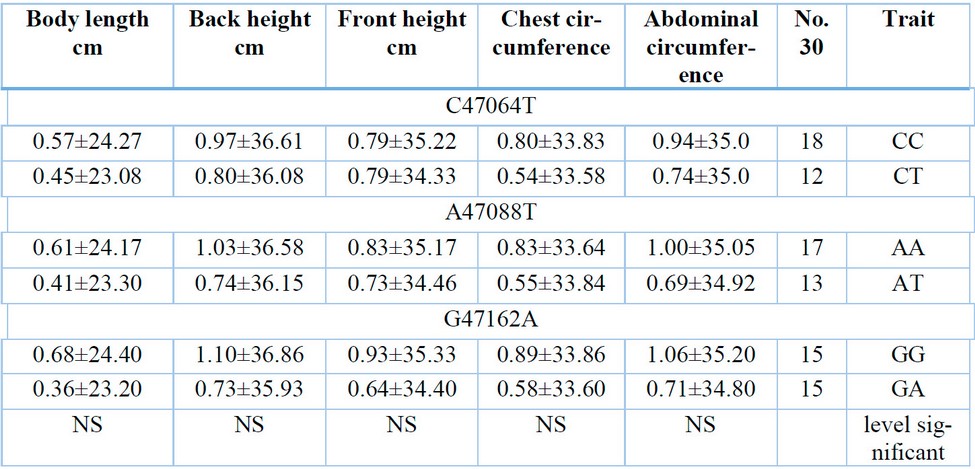
Table 5: Shows the effect of genotypes on the body dimensions of lambs at birth.
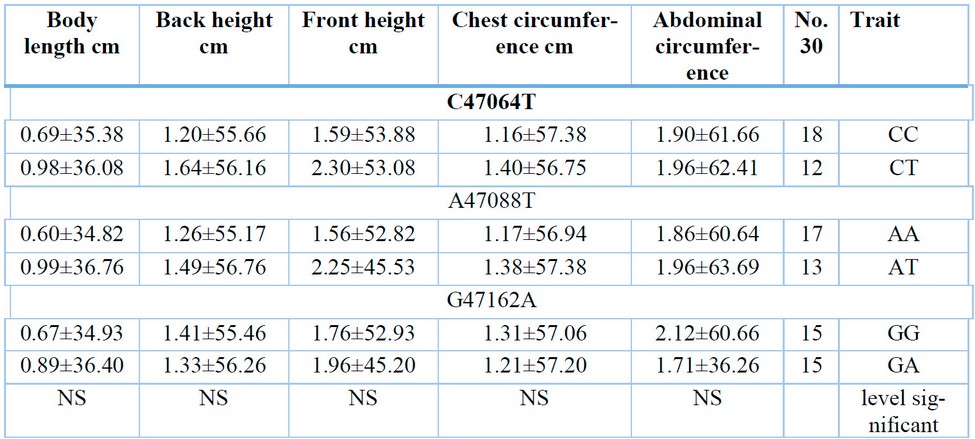
Table 6. Shows the effect of genotypes on the body dimensions of lambs when weaning.
Table (3) shows no significant effect of the point mutations C47064T, A47088T, and G47162A on daily and total milk production characteristics. The result of this study is consistent with what was reached by 13 in their study on sheep, as they did not find any significant effect of the genotypes. STAT5A gene in total milk production, while the result of the current study differs from what 14 found in their research on breeds of goats, as they found superiority of the CC genotype at the 6852C>T mutation site in total milk production.
The current study showed that there were no significant differences for the studied point mutations C47064T, A47088T, and G47162A in birth weight, weaning weight, and total weight gain, as shown in Table (4), and the result of this study agreed with what was reached by 13 in their study on sheep, as no A significant effect of the genotypes of the STAT5A gene was found on birth weight, weaning, and daily weight gain, while the results of this study agreed with part of what was reached by 15 in their study on male goats, as they did not find a significant effect of the genotypes at the mutation site (T11289C) in... Body weight: As for the location of the mutation A14320G, the genotype (GG) is significantly superior to the two combinations (AG, AA) in this trait, 6852C>T.
The current study showed in Table (5) that there were no significant differences for the studied point mutations C47064T, A47088T, and G47162A in the body dimensions of the lambs at birth. This study is partially consistent with what 15 reached in their study on male goats, as they did not find a significant effect of the genotypes. Site The mutation (T11289C) and (A14320G) in both body length, body height and shoulder width, and they found the superiority of the two genotypes CC (mutation site (T11289C)) and GG mutation site (A14320G) in the depth of the chest, and it partly agreed with what 16 found in their study on females. Goats aged 2-3 years found superiority of the mutant genotype (TT) in body height, chest circumference, and shoulder width.
It is clear from Table (6) that there are no significant differences for the studied point mutations in body dimensions at weaning. This result contradicts what 16 reached in his study on 2-3-year-old females of Chinese goats, the HNBG breed, for meat production. They found a significant superiority of the mutant genotype (TT) compared to the two genotypes (CC and TC) in chest circumference, body height, and butt width. They also found no significant effect of the genotypes of this gene in both body weight and chest depth.
CONCLUSIONS
The results of this study showed the presence of three mutations within the studied region of the seventh exon: the first mutation at the C47064T site resulting from the transformation of the base C to T, the second at the A47088T site resulting from the transformation of the base A to T, and the third mutation G47162A resulting from the transformation of the G base to A. The studied mutations had no significant effect on milk production and growth characteristics.
REFERENCES
1. Gipson, A. Terry. Recent advances in breeding and genetics for dairy Goats 2019. Asian-Australas J Anim Sci. 32(8): 1275–1283.
2. Raziye, I. and Güldehen, B. Associations between genetic variants of the POU1F1 gene and production traits in Saanen goats. 2019. Arch. Anim. Breed., 62, 249–255.
3. Oget, C., Servin, B. and Palhière, I. Genetic diversity analysis of French goat populations reveals selective sweeps involved in their differentiation. Anim Genet. 2019.; 50:54–63.
4. Alakilli, S. Y. M., Karima, F. Mahrous, Lamiaa, M. S. and Ekram, S A. Genetic polymorphism of five genes associated with growth traits in goat. 2012. African Journal of Biotechnology Vol. 11(82), pp. 14738- 14748.
5. Williams, J. L. The use of marker-assisted selection in animal breeding and biotechnology. 2005. Rev. Sci. Tech. 24(1): 379-91.
6. Chughtar N, Schimchowitsch S, Lebrun JJ, Ali S. Prolactin induces SHP-2 association with Stat5, nuclear translocation, and binding to the beta-casein gene promoter in mammary cells. 2012. J Biol Chem. 277:31107.
7. Brym, S. Kamiński, and Ruść A. New SSCP polymorphism within bovine STAT5A gene and its associations with milk per-formance traits in Black-and-White and Jersey cattle. 2004. Journal of Applied Genetics, vol. 45, pp. 445-452.
8. Wakao H, Gouilleux F and Groner. B . Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. 1994. EMBO J 13:2182–2191.
9. Aaronson, D. S. and Horvath, C. M. A road map for those who don't Know JAK-STAT. 2002. Science, 296(5573):1653-1655.
10. Cam, M. A.; M. Olfaz and E. Soydan. Body Measurements Reflect Body Weight and Carcass Yields in Kara Yaka Sheep. 2010. Asian J. Anim .Vet.Adv,(5):120-127.
11. SAS . SAS/STAT User's Guide for Personal Computers . 2012. Release 9.1 SAS Institute Inc. , Cary , N. C. , USA .
12. Duncan, D.D. Multiple range and multiple F-test Biometrics 1955 , 11: 1-42.
13. Abousoliman, I., Reyer, H., Oster, M., Muráni, E., Mourad, M., Abdel-Salam Rashed, M., & Wimmers, K. Analysis of candidate genes for growth and milk performance traits in the Egyptian Barki sheep. Animals, 2020. 10(2), 197.
14. An, X. P., Hou, J. X., Zhao, H. B., Bai, L., Peng, J. Y., Zhu, C. M., ... & Cao, B. Y. Polymorphism identification in goat DGAT1 and STAT5A genes and association with milk production traits. Czech J. Anim. Sci, 2013. 58(7), 321-327.
15. Xie, H. Q., Sun, Y. Y., Pan, D. X., Yang, Y. Q., Jiao, R. G., Gong, Y., ... & Liu, R. Y. Association analysis between polymorphism of STAT5a gene and growth traits in Chinese guizhou black goats. Pak. J. Agri. Sci2015., 52(4), 1119-1123.
16. Wu, X., Jia, W., Zhang, J., Li, X., Pan, C., Lei, C., & Lan, X. Determination of the novel genetic variants of goat STAT5A gene and their effects on body measurement traits in two Chinese native breeds. Small Ruminant Research, 2014. 121(2-3), 232-243.
Received: October 9th 2023/ Accepted: January 15th 2024 / Published:15 February 2024
Citation: Bashar A.; Hamza K. Relationship Polymorphism of STAT5A Gene in Performance Production of Local Goats. Revis Bionatura 2024; 1 (1) 57. http://dx.doi.org/10.21931/BJ/2024.01.01.57
Additional information Correspondence should be addressed to gabriela.rosero@espoch.edu.ec
Peer review information. Bionatura thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).

